Introduction to Creating Awesome Plots with ggplot2
 Plot Created with ggplot2 by Georgios Karamanis
Plot Created with ggplot2 by Georgios Karamanis
The package “ggplot2” has become the go-to package for making publication-quality plots. It is part of the tidyverse It can be complicated to use. It derived from the “grammar of graphics” concept that aims to have a consistent set of options to build a diverse set of simple or complex plots from data.
Data and packages
We will load ggplot2, which will automatically load a useful package of color palette, ‘RColorBrewer’. The packages ‘dplyr’, ‘forcats’ and ‘data.table’ are loaded since we will need to use some of their functions to prepare the data for plots.
The data set, “nass.hay” is from the ‘agridat’ package. It is historic data on hay production across U.S. states from 1909 to 2012 gathered by the National Agricultural Statistics Service. This script also creates a data object only containing data from Idaho, and a data object with hay production stats from 2012.
library(agridat) # loaded so we can access the following date set:
data("nass.hay")
str(nass.hay)
## 'data.frame': 5044 obs. of 4 variables:
## $ year : int 1909 1909 1909 1909 1909 1909 1909 1909 1909 1909 ...
## $ state: Factor w/ 49 levels "Alabama","Alaska",..: 1 3 4 5 6 7 8 9 10 11 ...
## $ acres: num 275000 98000 409000 2503000 1180000 ...
## $ yield: num 0.96 2.42 1.05 1.61 1.73 1.15 1.22 0.69 0.86 2.16 ...
head(nass.hay)
## year state acres yield
## 1 1909 Alabama 275000 0.96
## 2 1909 Arizona 98000 2.42
## 3 1909 Arkansas 409000 1.05
## 4 1909 California 2503000 1.61
## 5 1909 Colorado 1180000 1.73
## 6 1909 Connecticut 387000 1.15
hay_idaho <- filter(nass.hay, state == "Idaho")
hay2012 <- filter(nass.hay, year == 2012)
hay_pnw <- filter(nass.hay, state %in% c("Oregon", "Washington", "Montana","Idaho")) %>%
mutate(state = fct_drop(state))
# fct_drop() command run so extra factor levels in "state" that are not longer represented in "hay_pnw" are dropped
Basics of ggplot
A ggplot needs 3 parts:
- data
- aesthetics which connect or map the data to the geom
- a type of plot, or geom, to implement
Note:
- ggplot2 uses the
+notation in their plots (see below). I think there is some regret over this style decision, but it is now an accepted convention! - ggplot2 also uses non-standard evaluation, so, often column names do not need to be quoted
- common mapping aesthetics:
- x (the x variable)
- y (the y variable)
- col (color for lines, points, outlines)
- fill (color when something has a fill color such as a boxplot)
- group (when a plotted option should be grouped, then plotted)
build a simple plot
Let’s plot Yield over time for Idaho hay production
Step 1: Add the data
ggplot(hay_idaho)

(not much to see here - we have not specified the aesthetics or the geom)
Step 2: Add aesthetics
ggplot(hay_idaho, mapping = aes(x = year, y = yield))
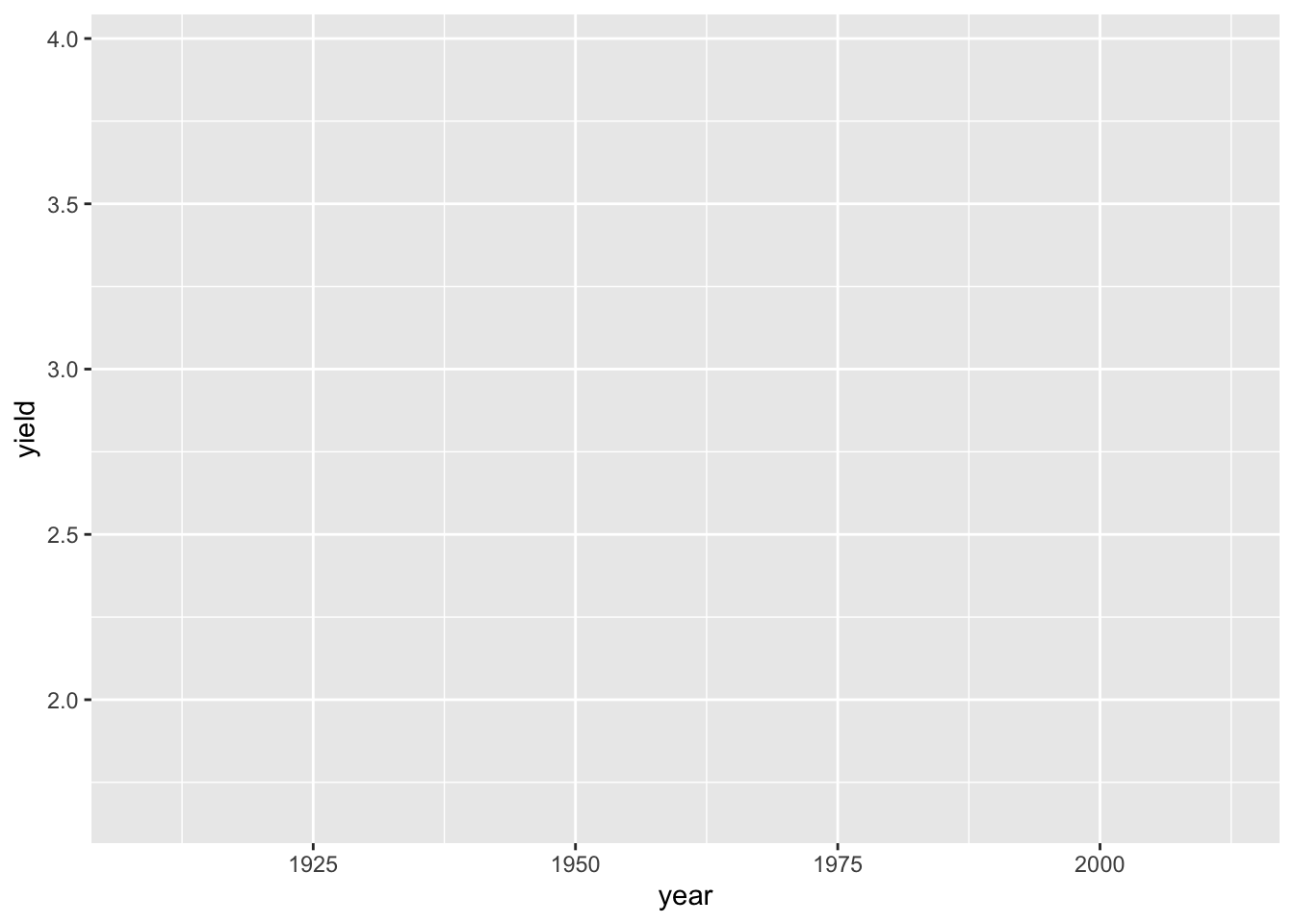
This step mapped “year” and “yield” from the hay_idaho data set to the x and y axes, respectively. Nothing is plotted because we have not specified what type of plot geom we want.
Step 3: Add a geom
(explicit mapping command dropped - use a positional argument instead)
ggplot(hay_idaho, aes(x = year, y = yield)) +
geom_line()
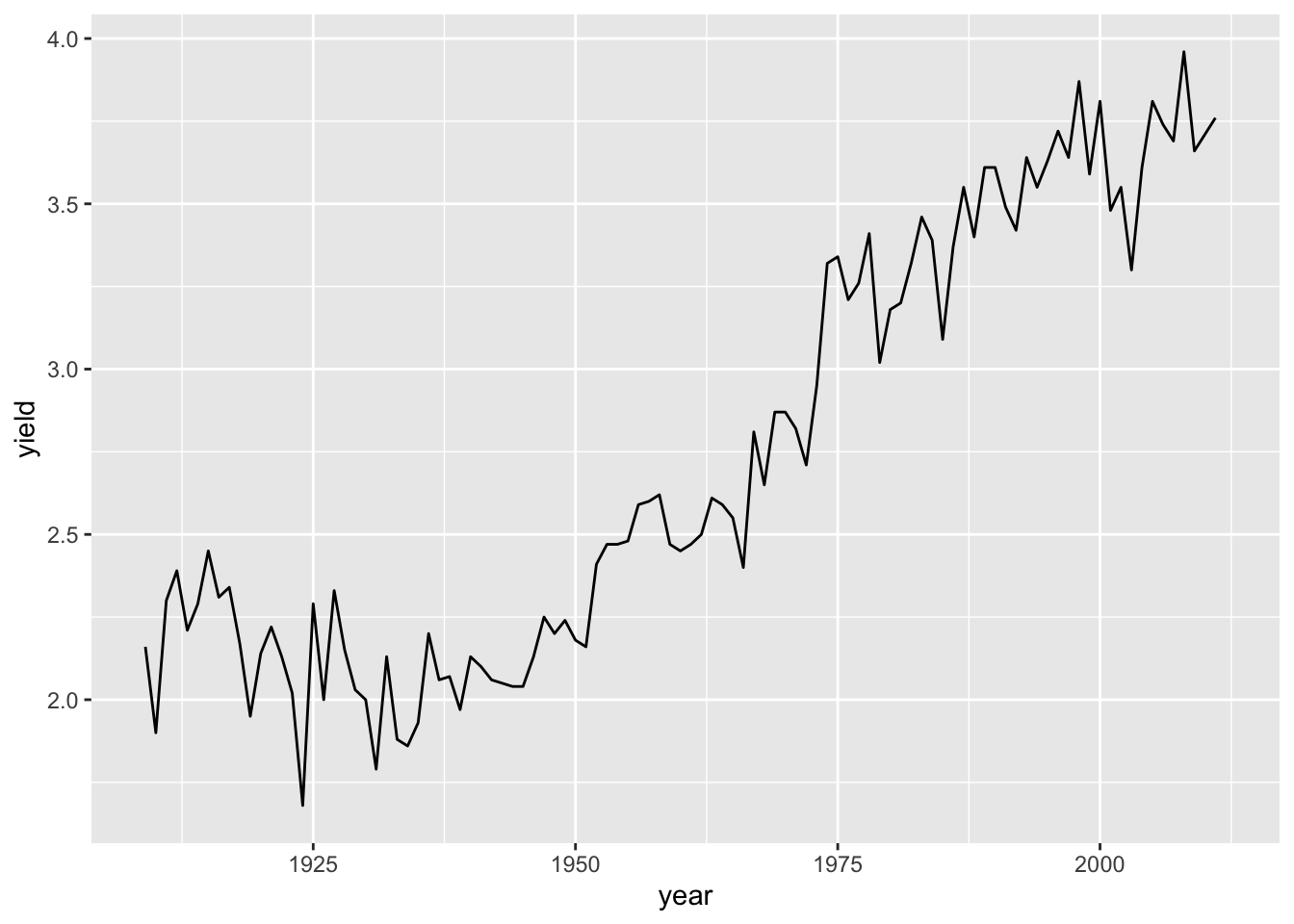 Question: What happens if we use
Question: What happens if we use %>% instead of +?
Common plot improvements
plot title and axis labels
ggplot(hay_idaho, aes(x = year, y = yield)) +
geom_line() +
ggtitle("Total historic hay yields in Idaho") +
xlab("Year") +
ylab("yield (1000s of tons)")
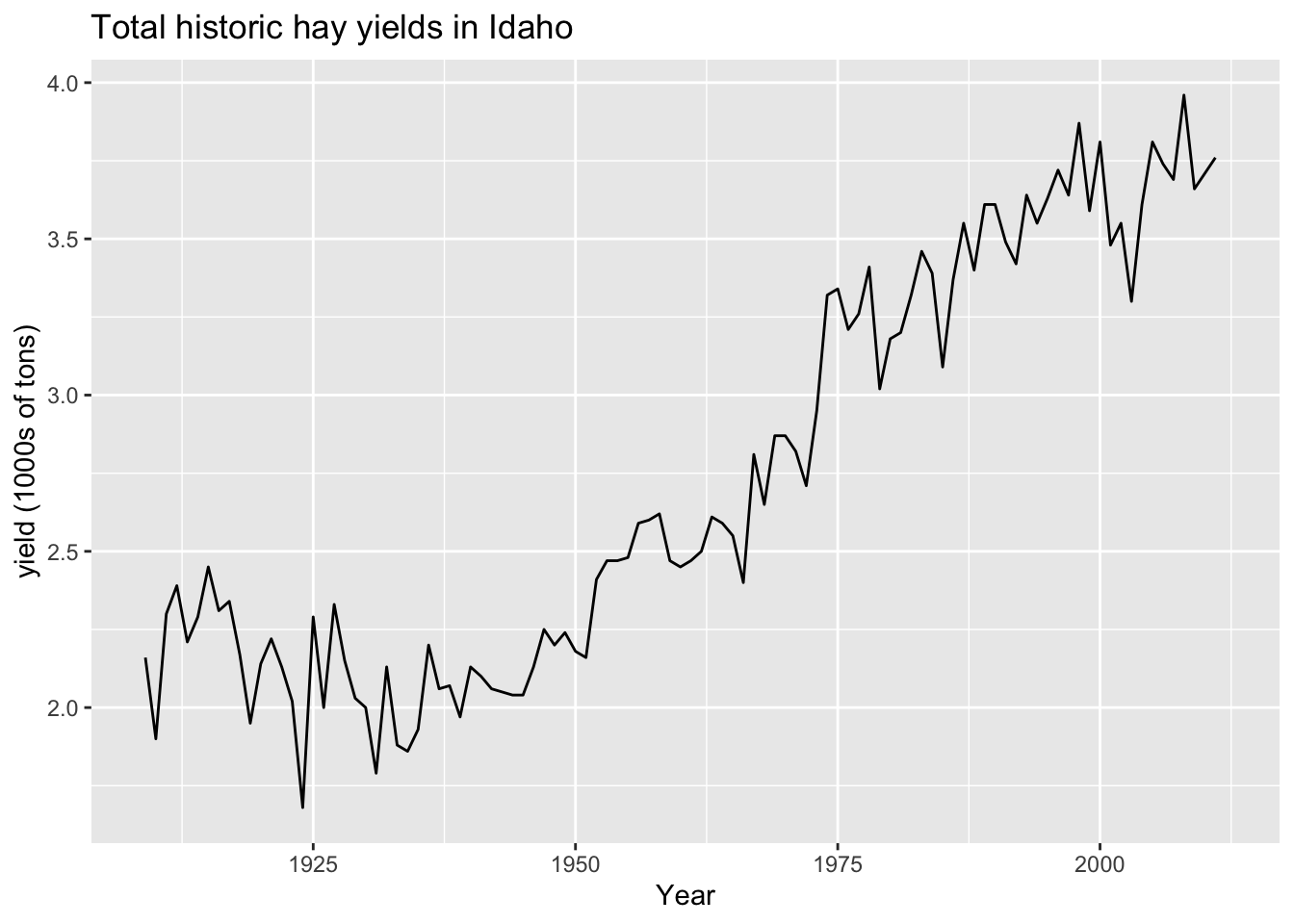
Change line settings
(and other geom arguments)
ggplot(hay_idaho, aes(x = year, y = yield)) +
geom_line(color = "turquoise", size = 1.5, linetype = 1) +
ggtitle("Total historic hay yields in Idaho") +
xlab("Year") +
ylab("yield (1000's of tons)")
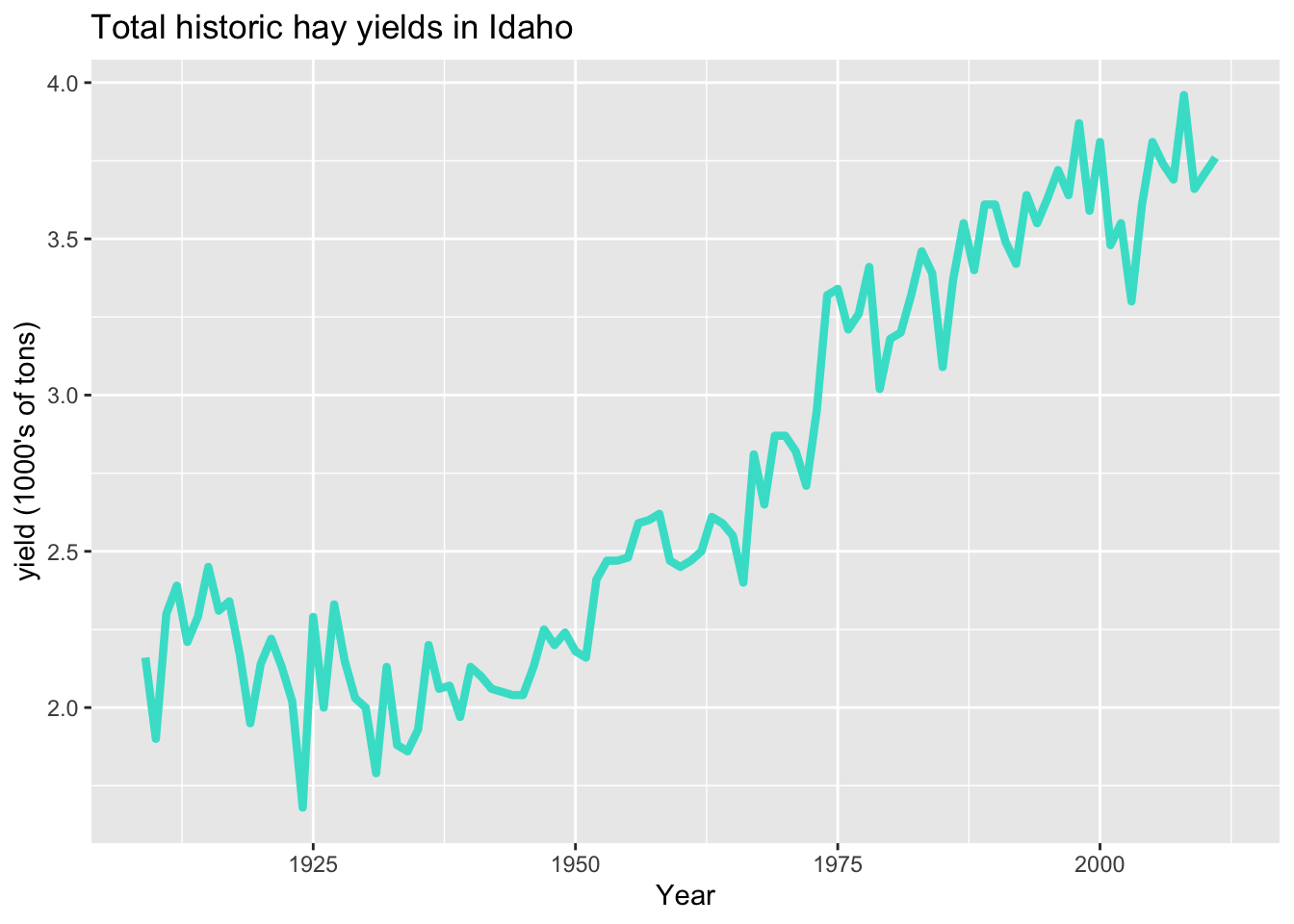
The default line type is “1”, which is a plain line. There are other types:

Other common geoms:
points
ggplot(hay_idaho, aes(x = year, y = yield)) +
geom_point(shape = 2, col = "navy") +
ggtitle("Total historic hay yields in Idaho") +
xlab("Year") +
ylab("yield (1000s of tons)")
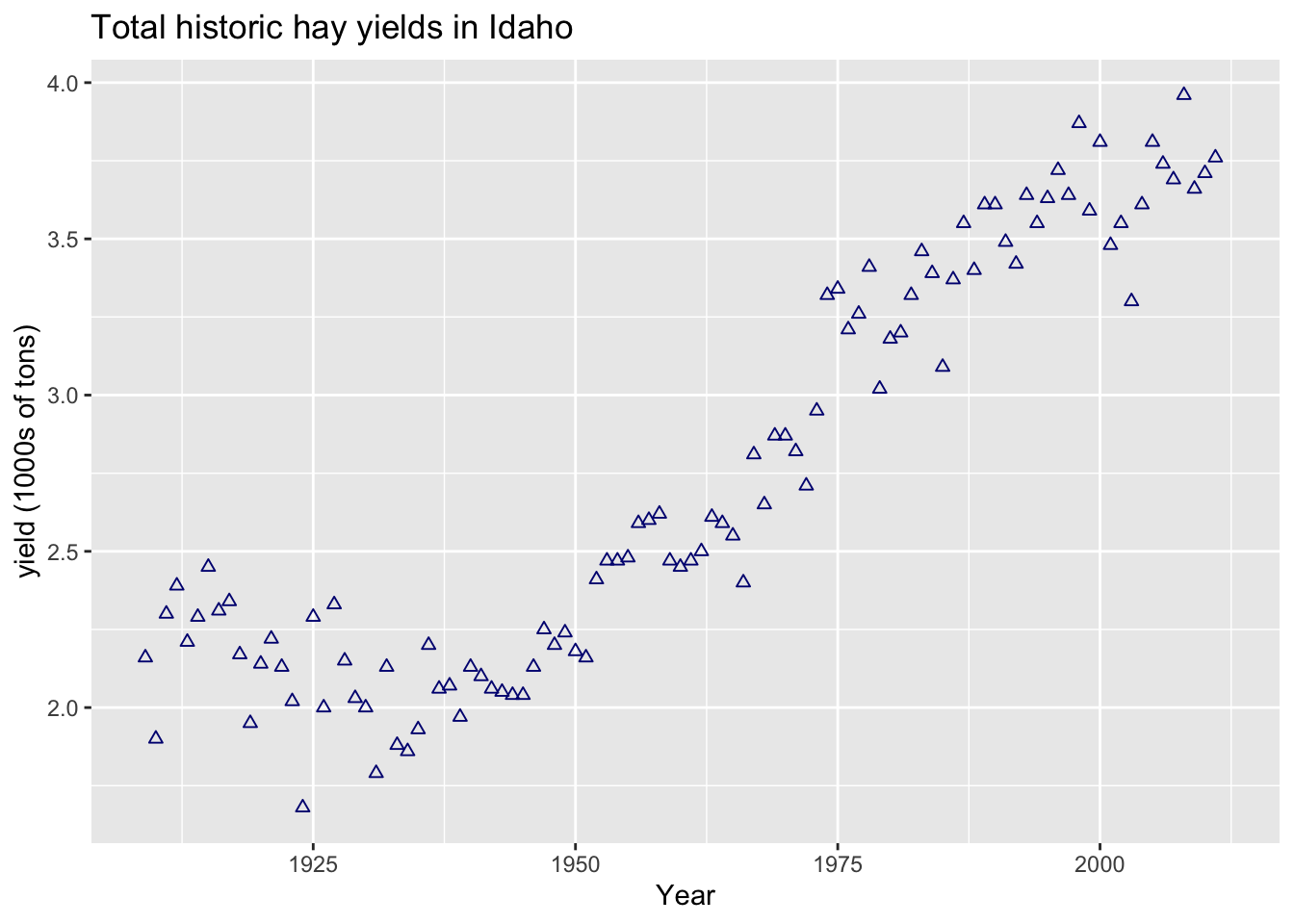
There are many options for the type of plotting character (“pch” in base R plotting and “shape” in ggplot):

The default shape is “1” (the circle). Plotting symbols 21 - 25 have outline color and fill attributes, while the other symbols only take a single color argument.
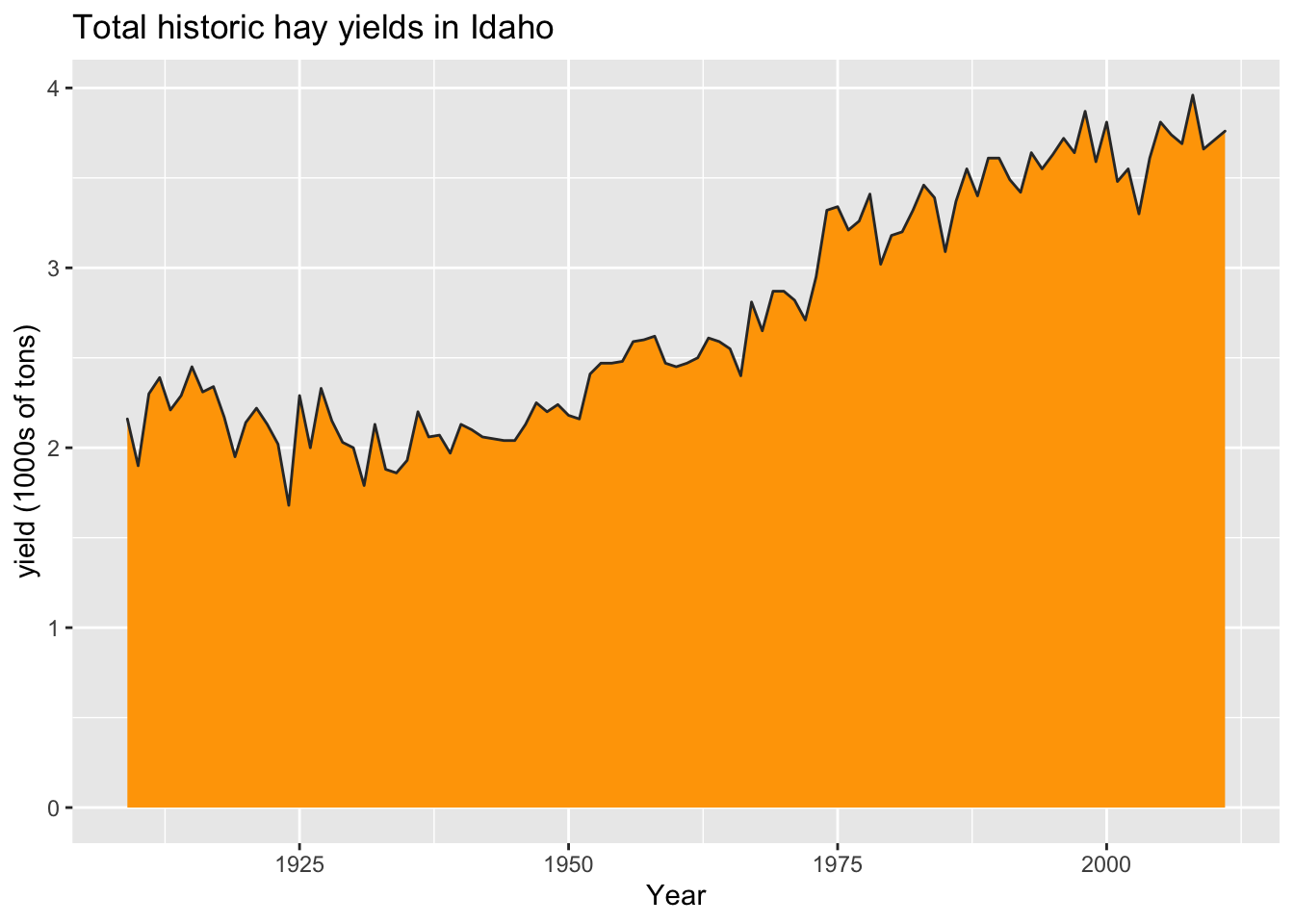
area
ggplot(hay_idaho, aes(x = year, y = yield)) +
geom_area(fill = "orange", col = "gray20") +
ggtitle("Total historic hay yields in Idaho") +
xlab("Year") +
ylab("yield (1000s of tons)")
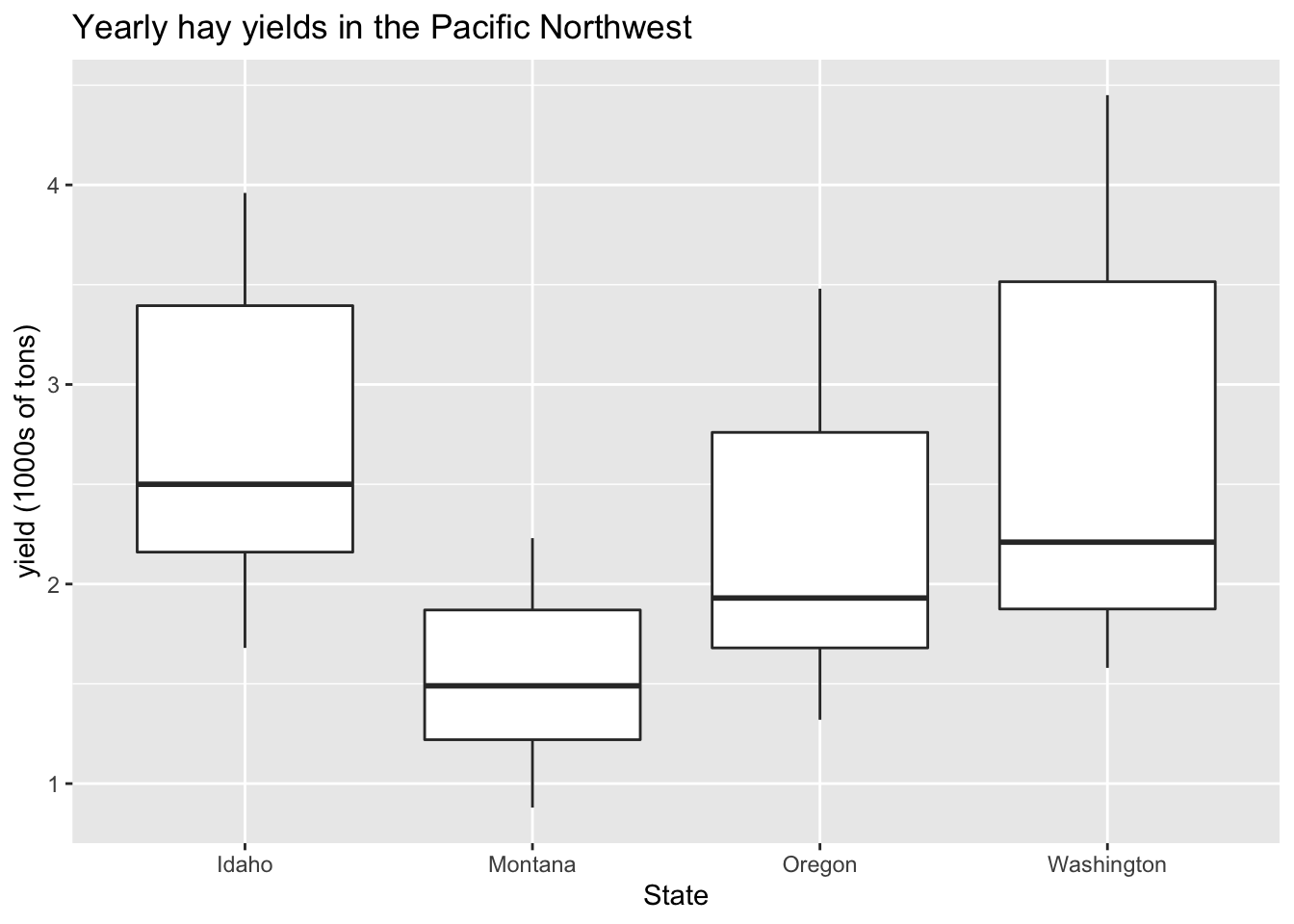
boxplot
ggplot(hay_pnw, aes(x = state, y = yield)) +
geom_boxplot() +
ggtitle("Yearly hay yields in the Pacific Northwest") +
xlab("State") +
ylab("yield (1000s of tons)")
Color all boxplots the same:
ggplot(hay_pnw, aes(x = state, y = yield)) +
geom_boxplot(fill = "yellow") +
ggtitle("Yearly hay yields in the Pacific Northwest") +
xlab("State") +
ylab("yield (1000s of tons)")
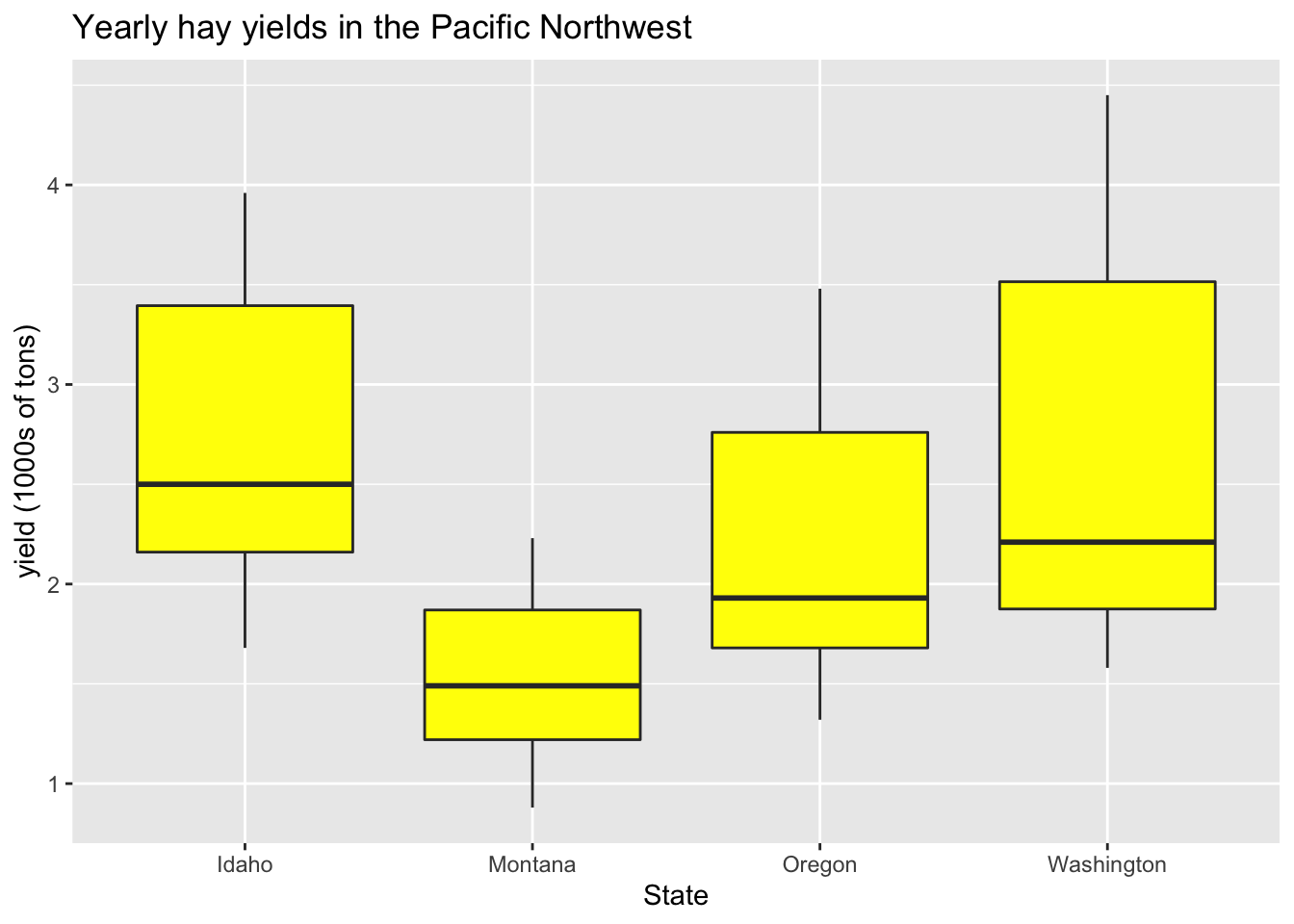
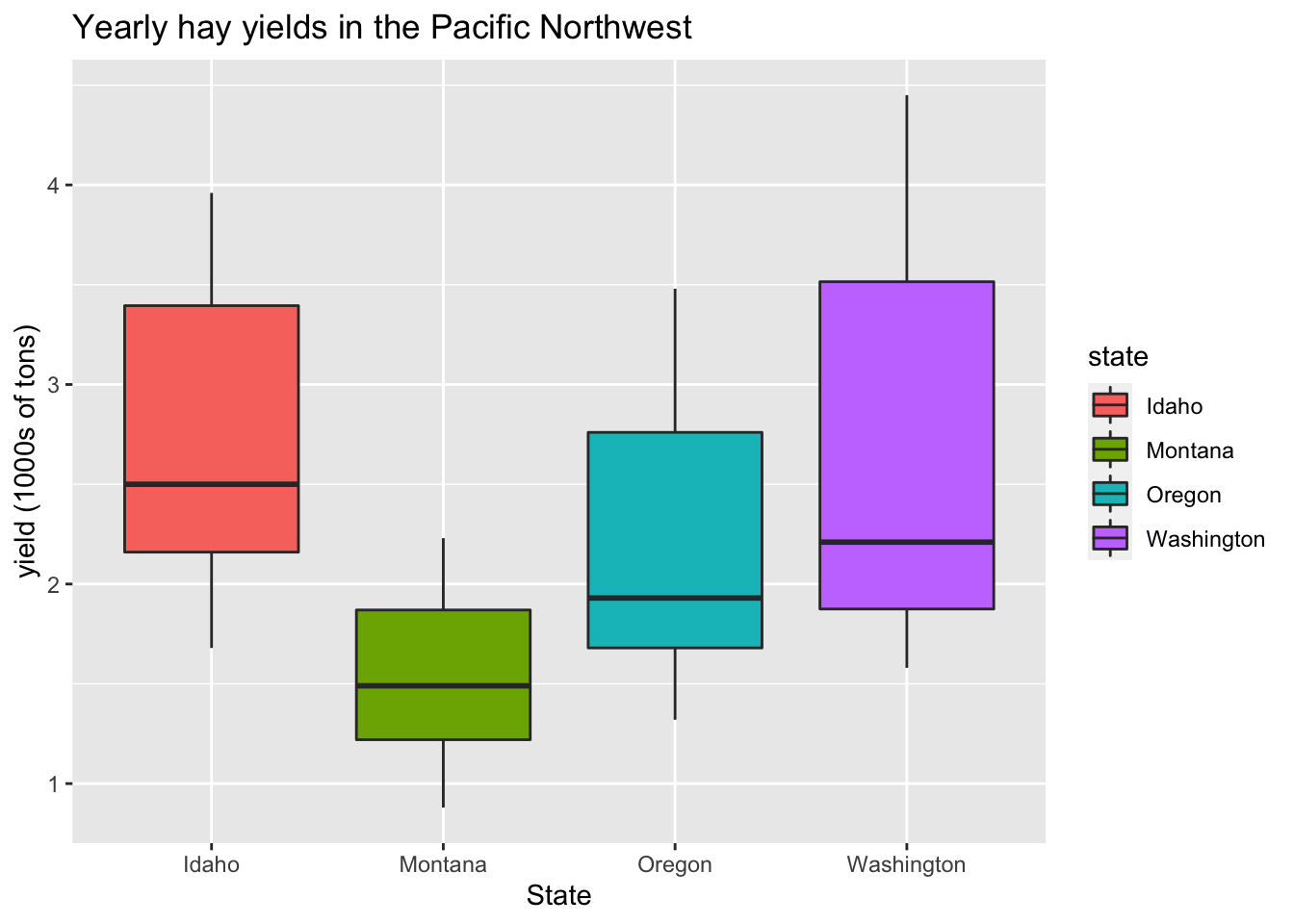
Color boxplots by state:
(This requires an aes statement.)
ggplot(hay_pnw, aes(x = state, y = yield, fill = state)) +
geom_boxplot() +
ggtitle("Yearly hay yields in the Pacific Northwest") +
xlab("State") +
ylab("yield (1000s of tons)")
 Question: What happens if we specify “color = state” instead of “fill = state”?
Question: What happens if we specify “color = state” instead of “fill = state”?
histograms and density plots:
ggplot(hay_idaho, aes(x = yield)) +
geom_histogram(fill = "gray70", col = "black", bins = 20)
## Warning: Removed 1 rows containing non-finite values (stat_bin).
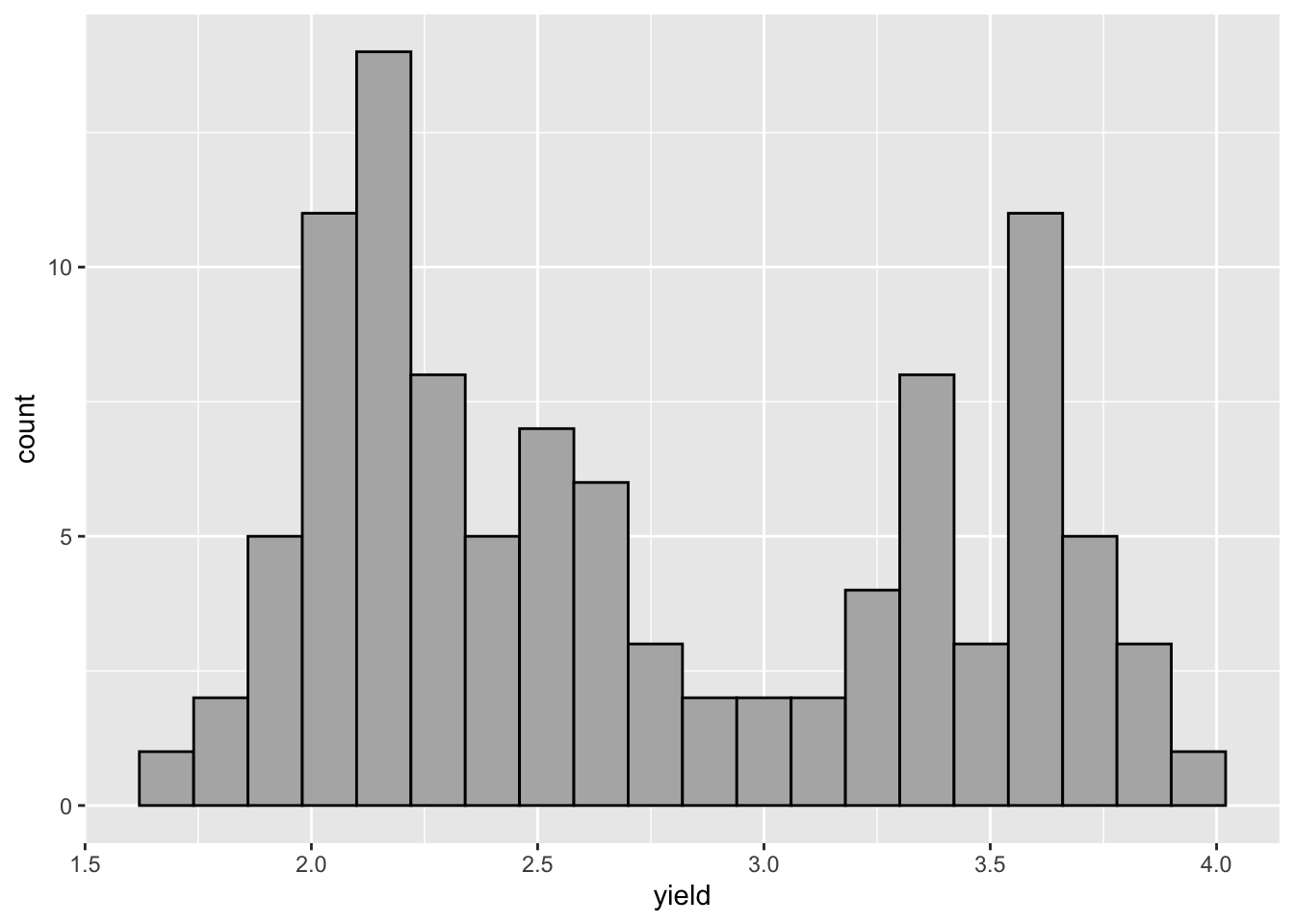
Unlike in base R plotting, creating a density plot and shading the are under the curve is easy.
ggplot(hay_pnw, aes(x = yield, fill = state)) +
geom_density()
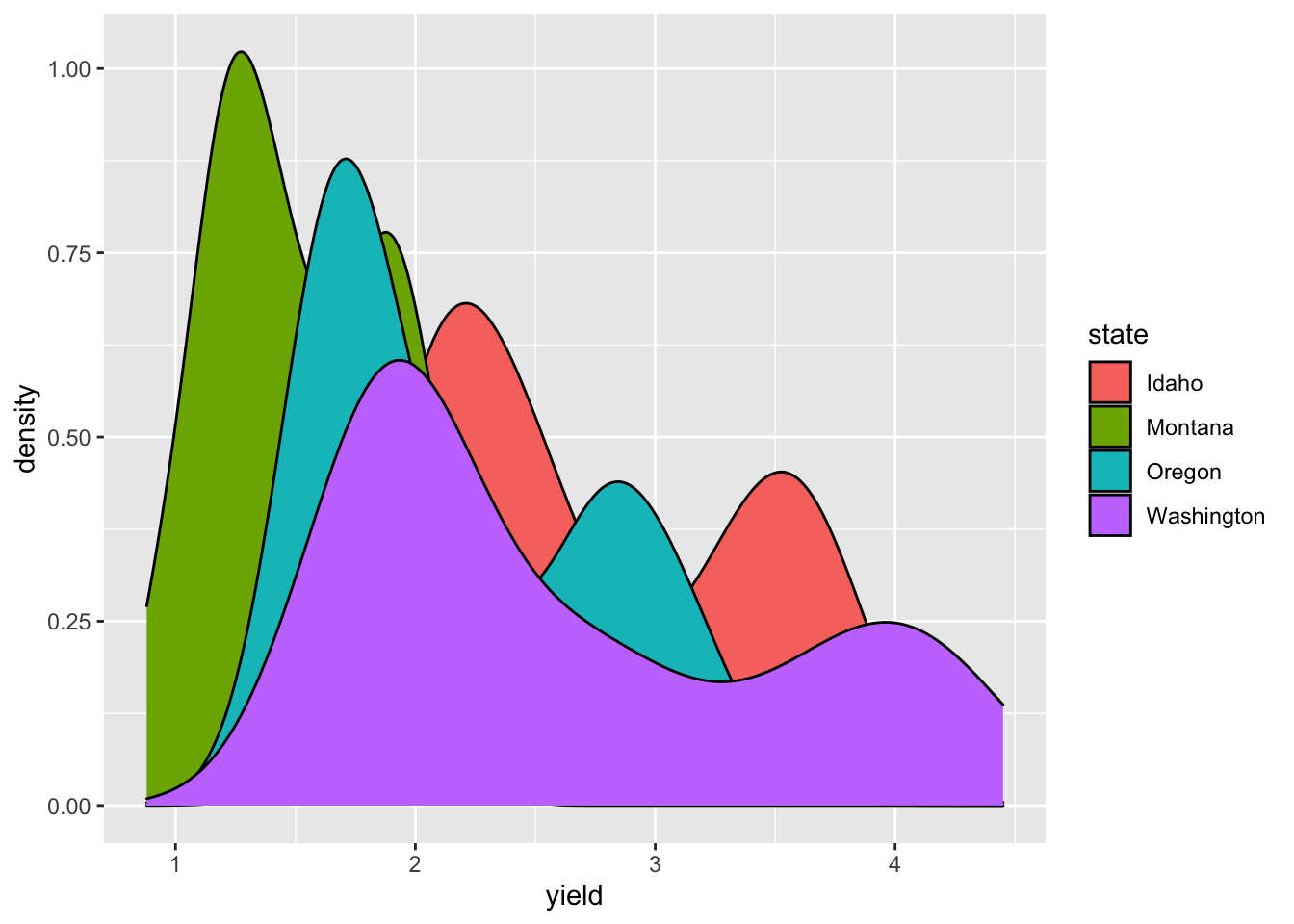
In this plot, it is hard to see what is going on since the each distribution covers up the one plotted before it (so we can’t really see Idaho, but Washington’s hay yield distribution is clear). The attribute “alpha” can be used to add a transparency to colors. The code is alpha = n where n is the proportion of the color that is shown (0 is completely transparent and 1 is no transparency).
ggplot(hay_pnw, aes(x = yield, fill = state)) +
geom_density(alpha = 0.5)
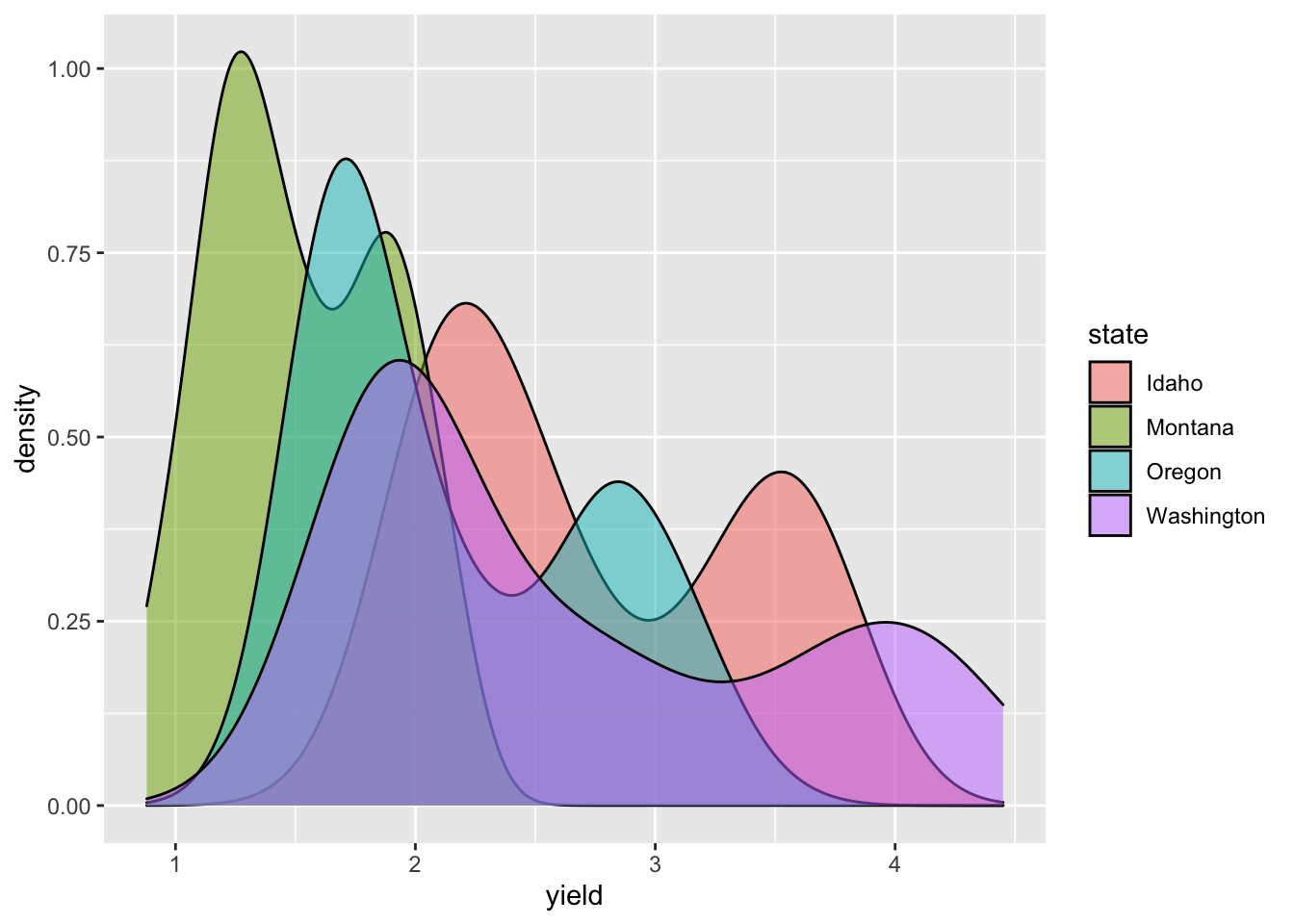
Combining geoms
Geoms are best thought of as plotting layers. Additional geoms can be added to a single plot, adding additional layers of information. Each geom is added to the plot in the order it is specified in the ggplot statement.
Adding an overall of points over boxplots is a good way to summarise the empirical distribution of a large number of data points:
ggplot(hay_pnw, aes(x = state, y = yield, fill = state)) +
geom_boxplot() +
geom_point() +
ggtitle("Yearly hay yields in the Pacific Northwest") +
xlab("State") +
ylab("yield (1000s of tons)")
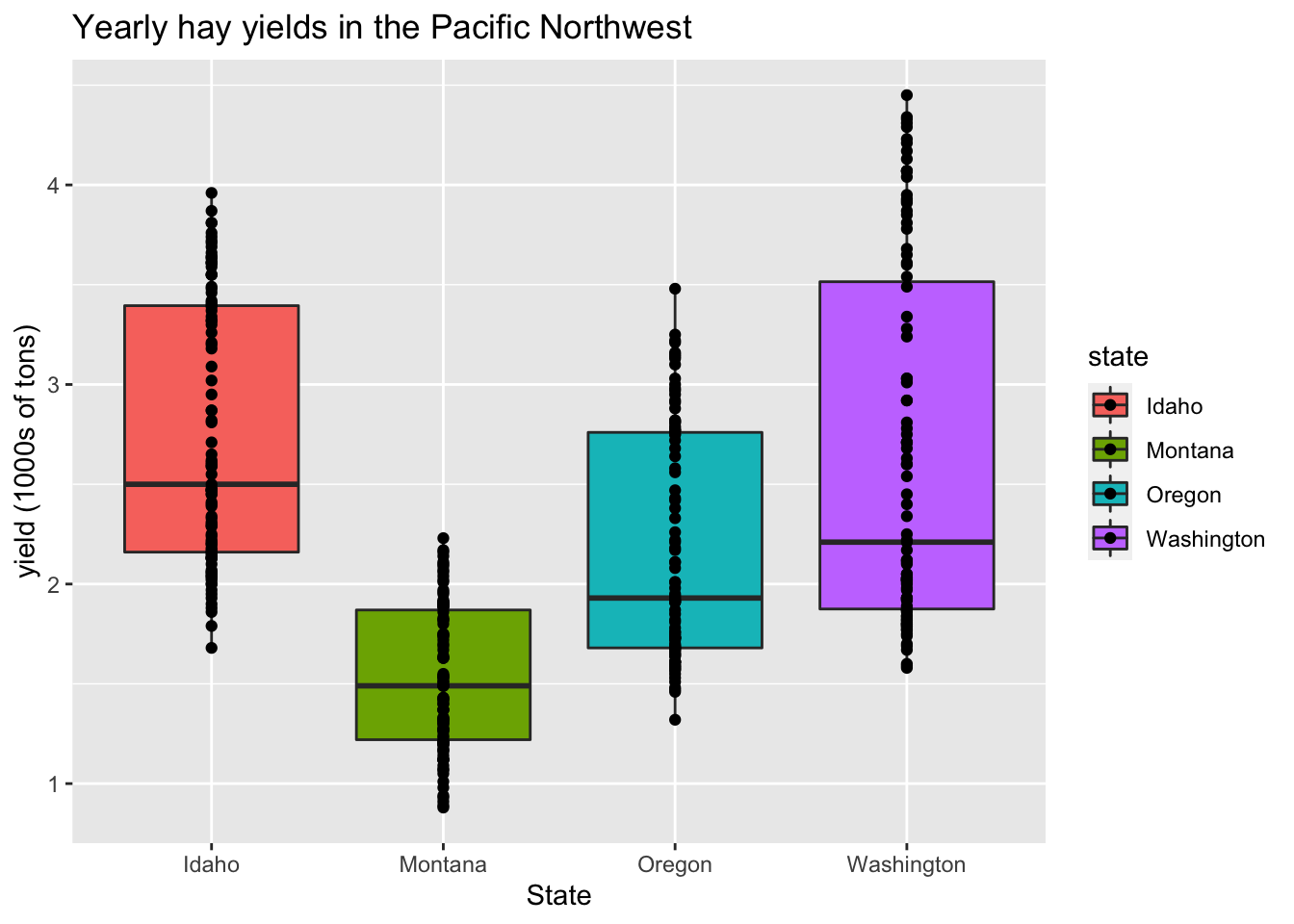
In this example, it is likely there is over-plotting of data since each state has 104 data point associated with it. The geom geom_jitter() can be used to jiggle overlapping points. Points can be jittered in the vertical and horizontal direction. In this case, jittering in the horizontal direction will result in no change in the interpretation since the y-axis indicates actual value associated with each point.
ggplot(hay_pnw, aes(x = state, y = yield, fill = state)) +
geom_boxplot() +
geom_jitter(width = 0.3, height = 0) +
ggtitle("Yearly hay yields in the Pacific Northwest") +
xlab("State") +
ylab("yield (1000s of tons)")
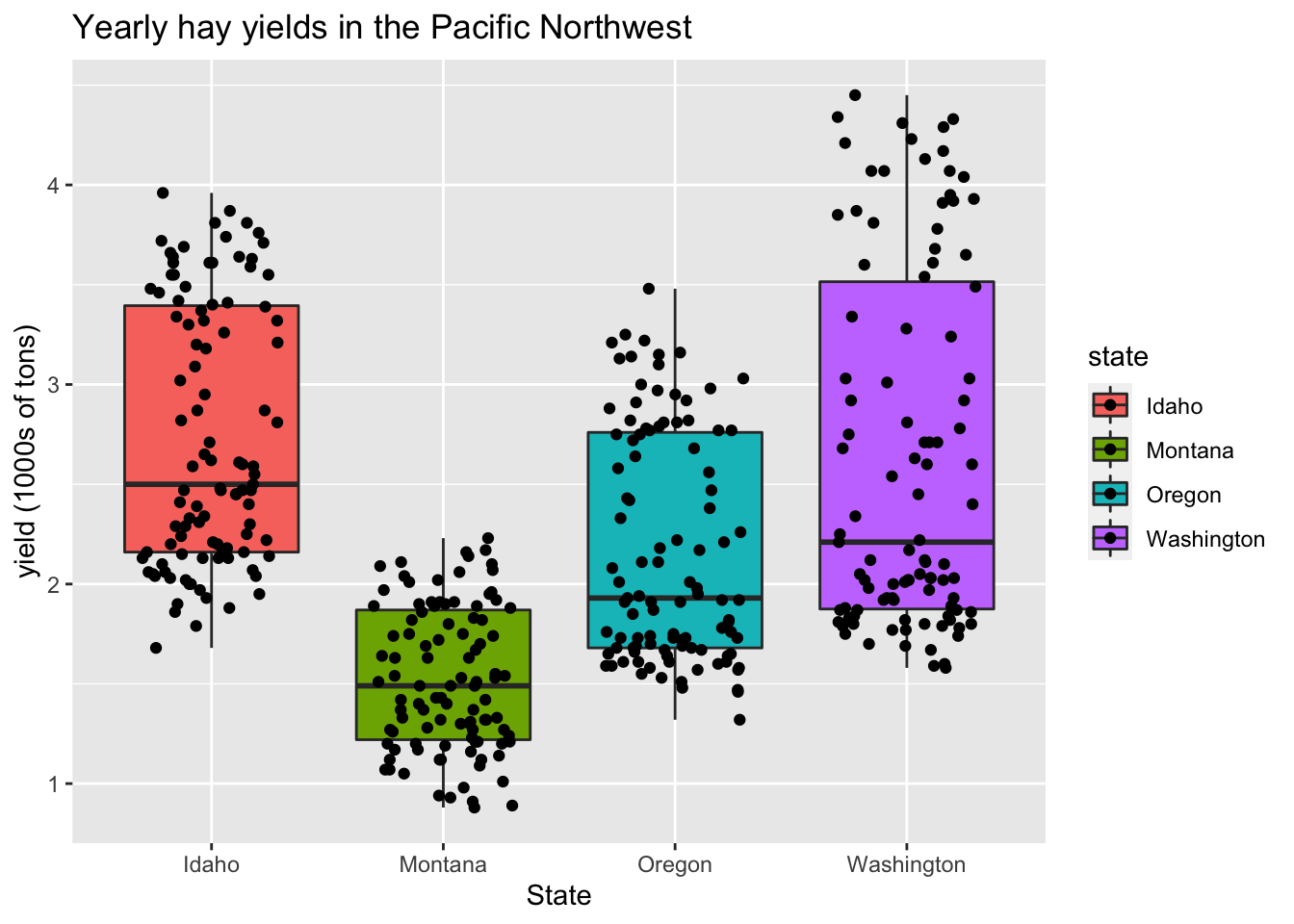
Other common plotting options
Faceting
Sometimes, it is useful to create a set of similar plots with the same scaling. Facets are useful for this purpose.
For an example, below is a plot of the distributions from above using facet_grid().
ggplot(hay_pnw, aes(x = yield)) +
geom_density(col = "blue", fill = "dodgerblue", alpha = 0.6) +
facet_grid(. ~ state) # for a horizontal facet
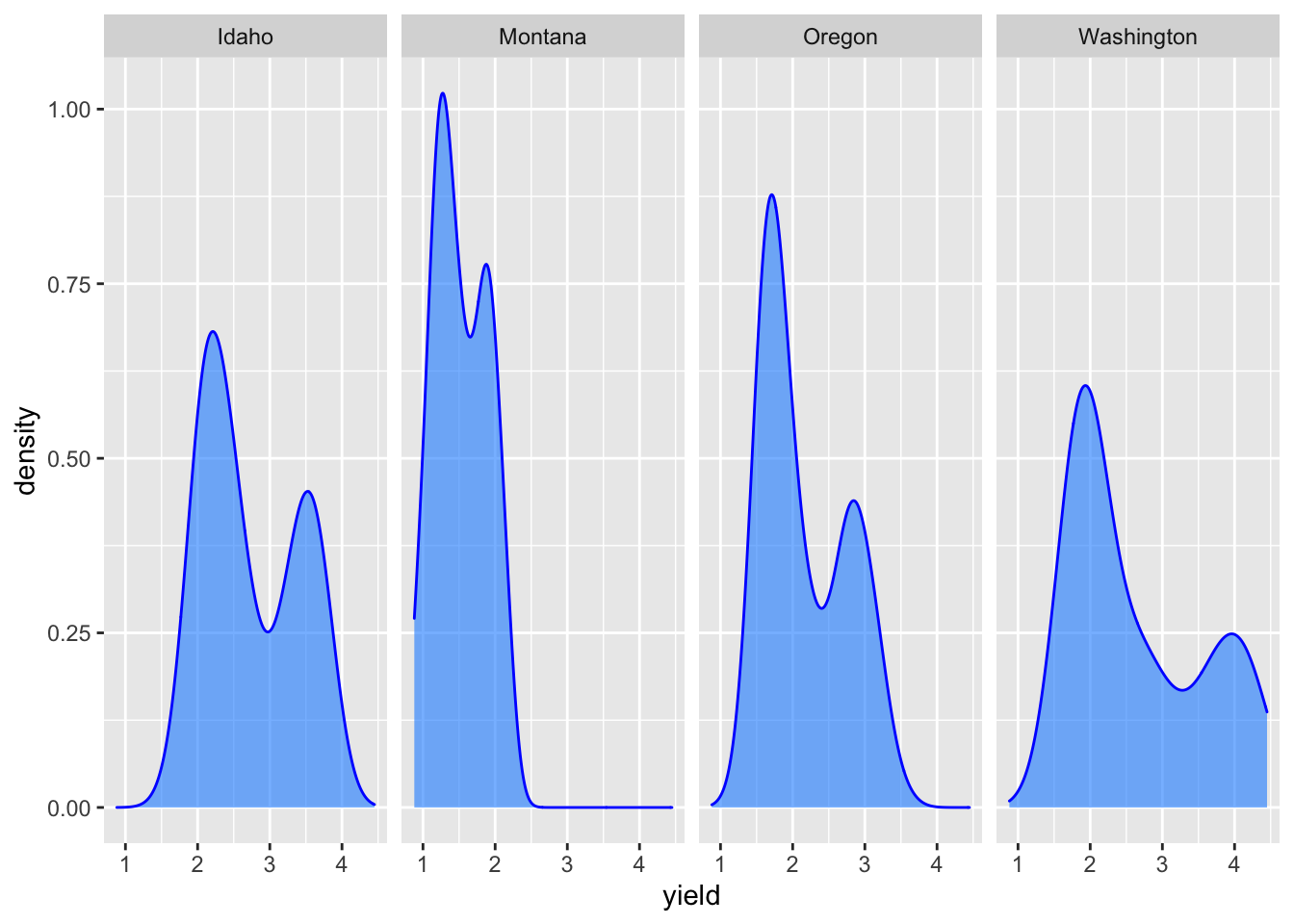
ggplot(hay_pnw, aes(x = yield)) +
geom_density(col = "darkgreen", fill = "springgreen3", alpha = 0.6) +
facet_grid(state ~ .) # for a vertical facet
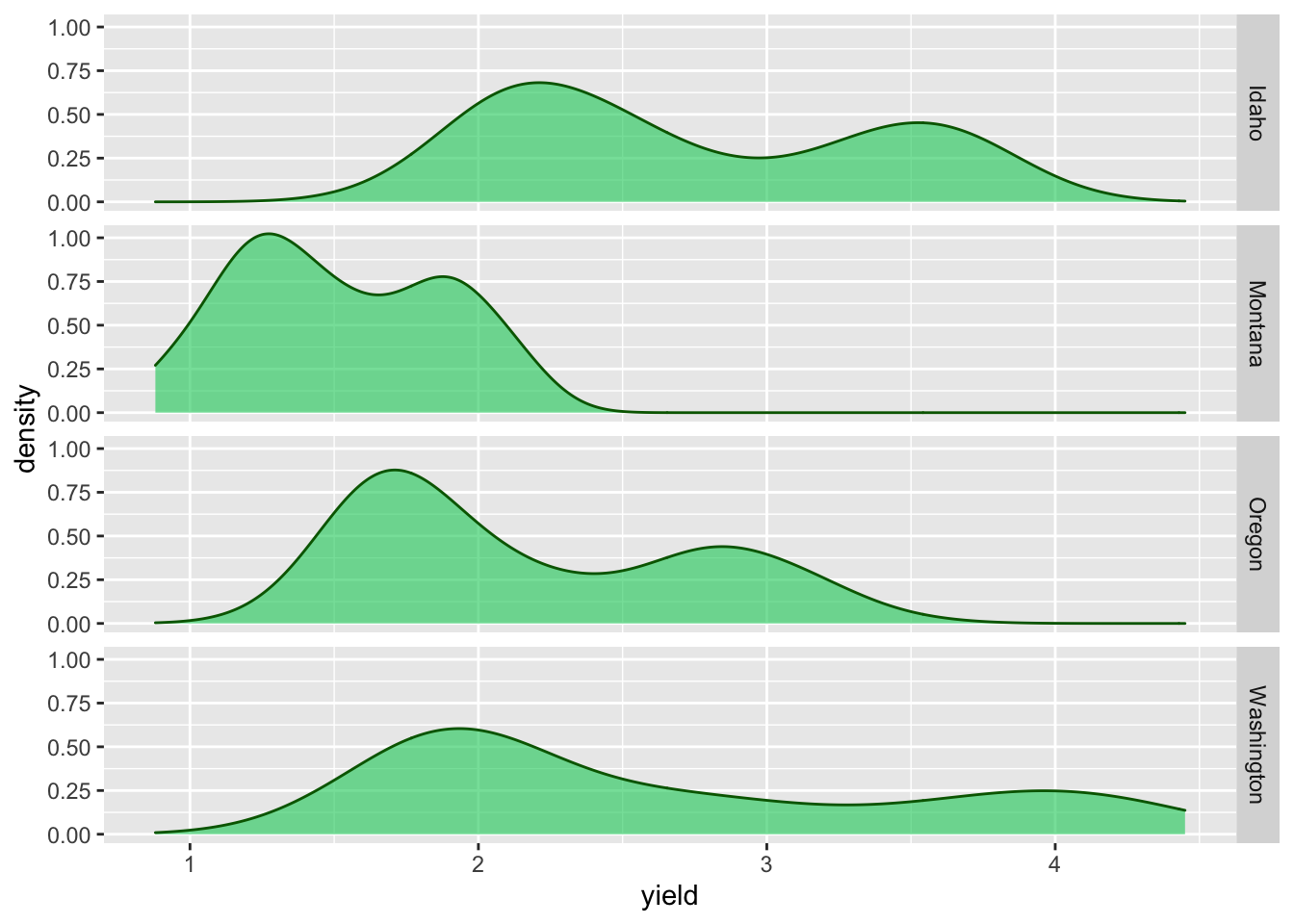
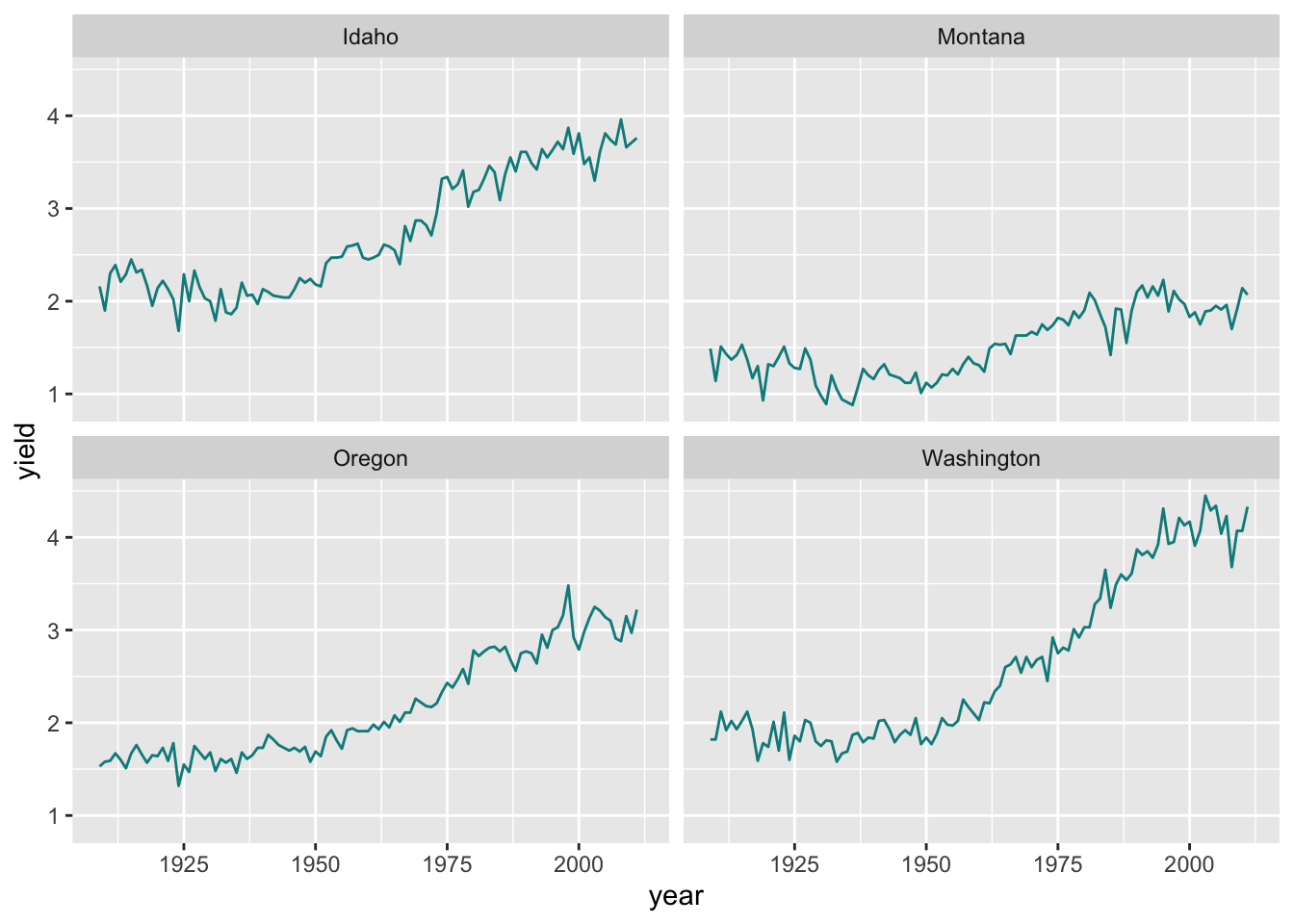
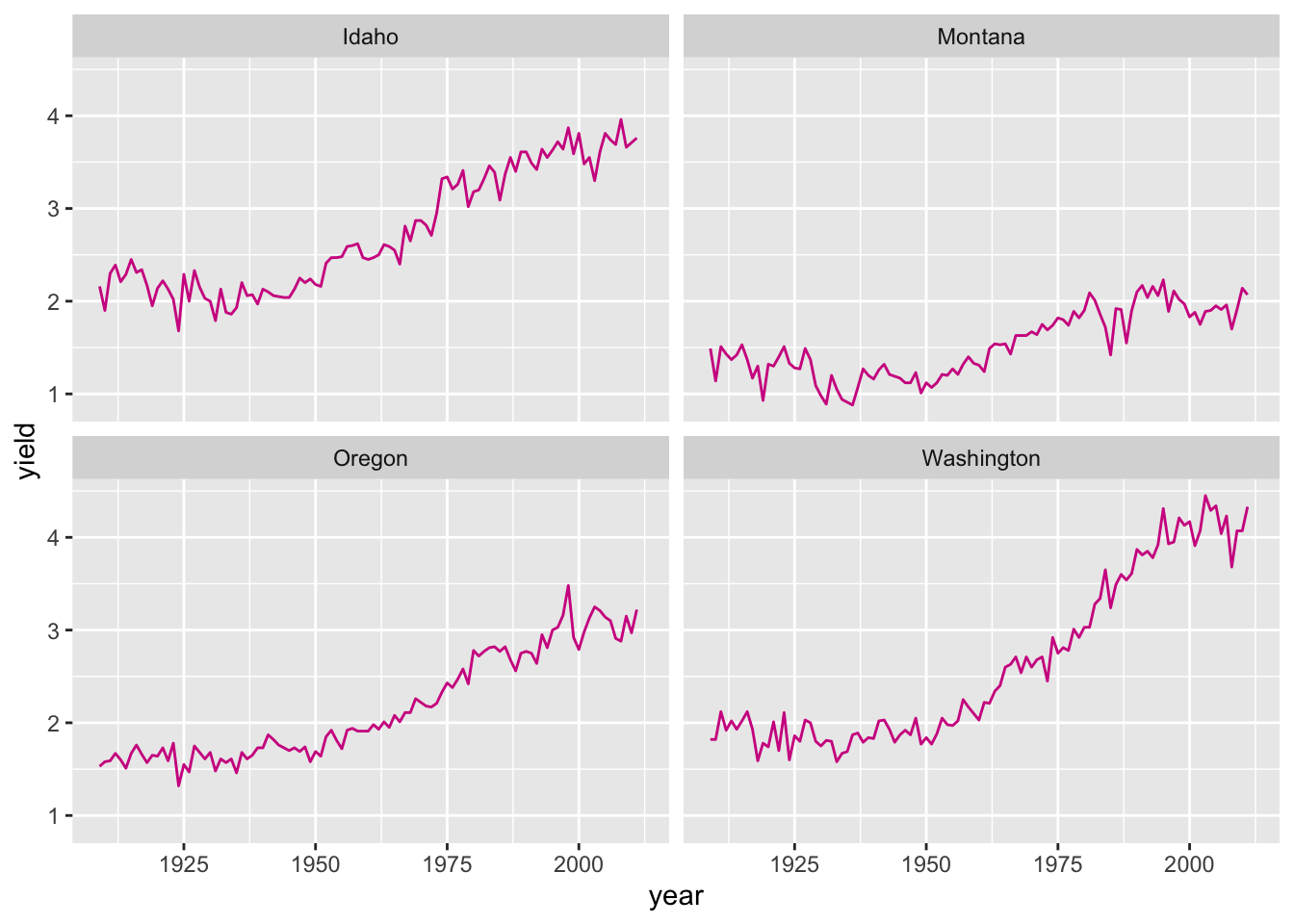
Faceting can also be done be used to create a grid of plots with facet_wrap():
ggplot(hay_pnw) +
geom_line(aes(x = year, y = yield), col = "darkcyan") +
facet_wrap(. ~ state, nrow = 2, ncol = 2)
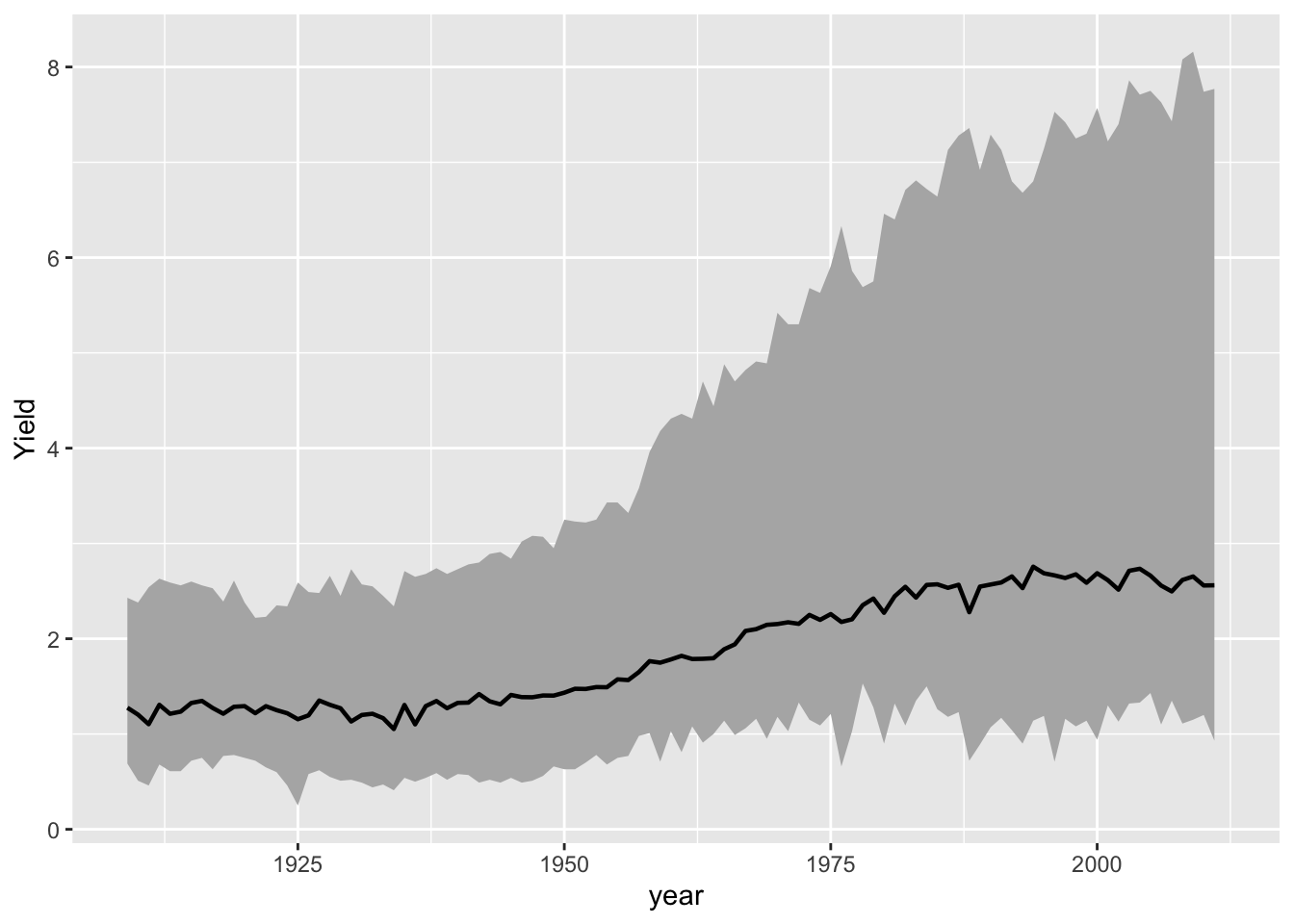
Representing error information
Example: create a band indicates the minimum and maximum range of values
hay_yearly <- nass.hay %>% group_by(year) %>%
summarise(Yield = mean(yield), yield_min = min(yield), yield_max = max(yield))
ggplot(hay_yearly, aes(x = year)) +
geom_ribbon(aes(ymin = yield_min, ymax = yield_max), fill = "gray70") +
geom_line(aes(y = Yield), size = 0.8)
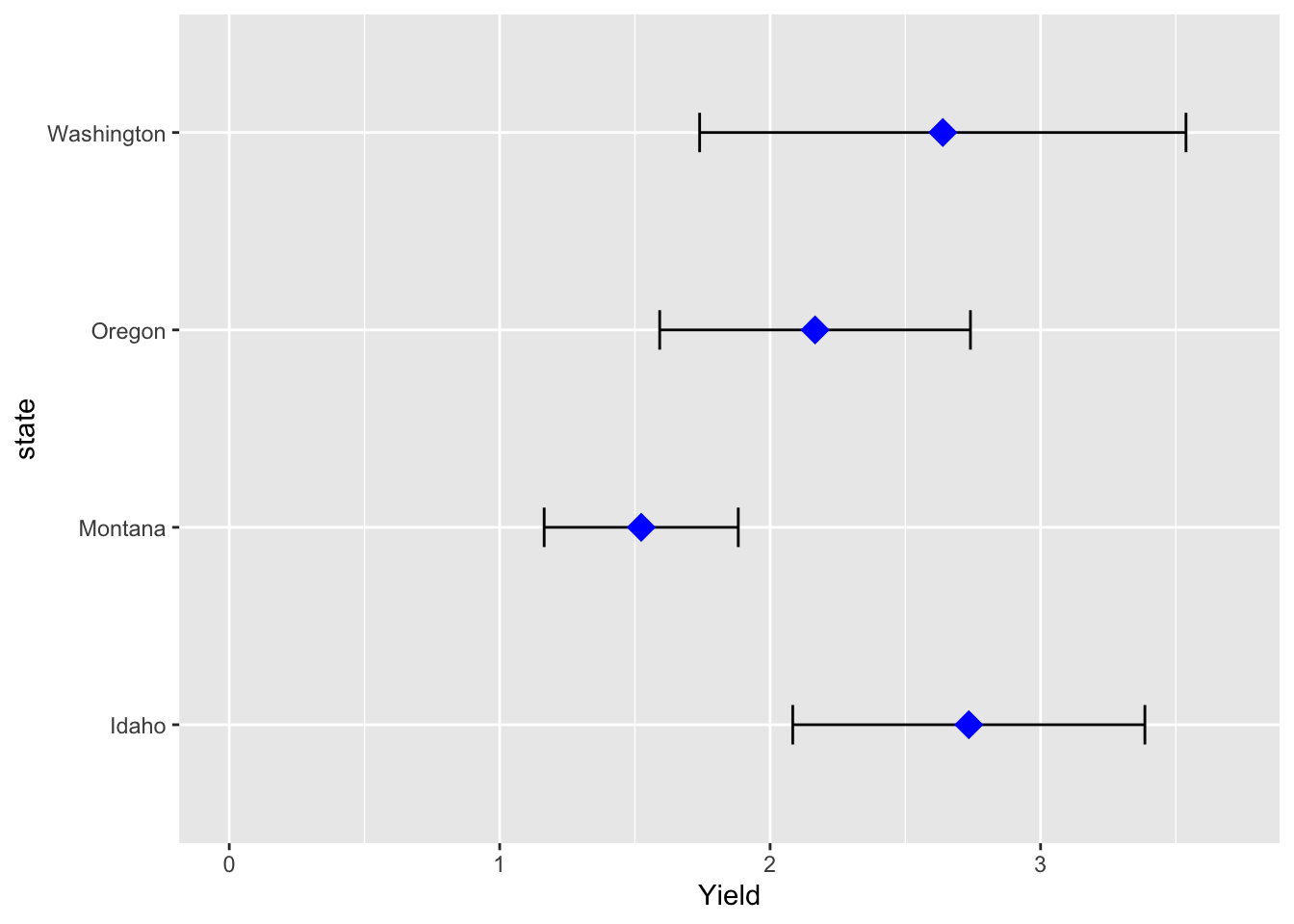
Example: add error bars
Two other plot adjustments included: * indicate the range of numbers that the y-axis should span * flip the x and y axes
hay_pnw_summary <- hay_pnw %>% group_by(state) %>%
summarise(Yield = mean(yield, na.rm = T), yield_sd = sd(yield, na.rm = T))
ggplot(hay_pnw_summary, aes(x = state)) +
geom_errorbar(aes(ymin = Yield - yield_sd, ymax = Yield + yield_sd), width = 0.2) +
geom_point(aes(y = Yield), shape = 18, size = 5, col = "blue") +
ylim(c(0, 3.7)) +
coord_flip()
Add regression lines to a plot
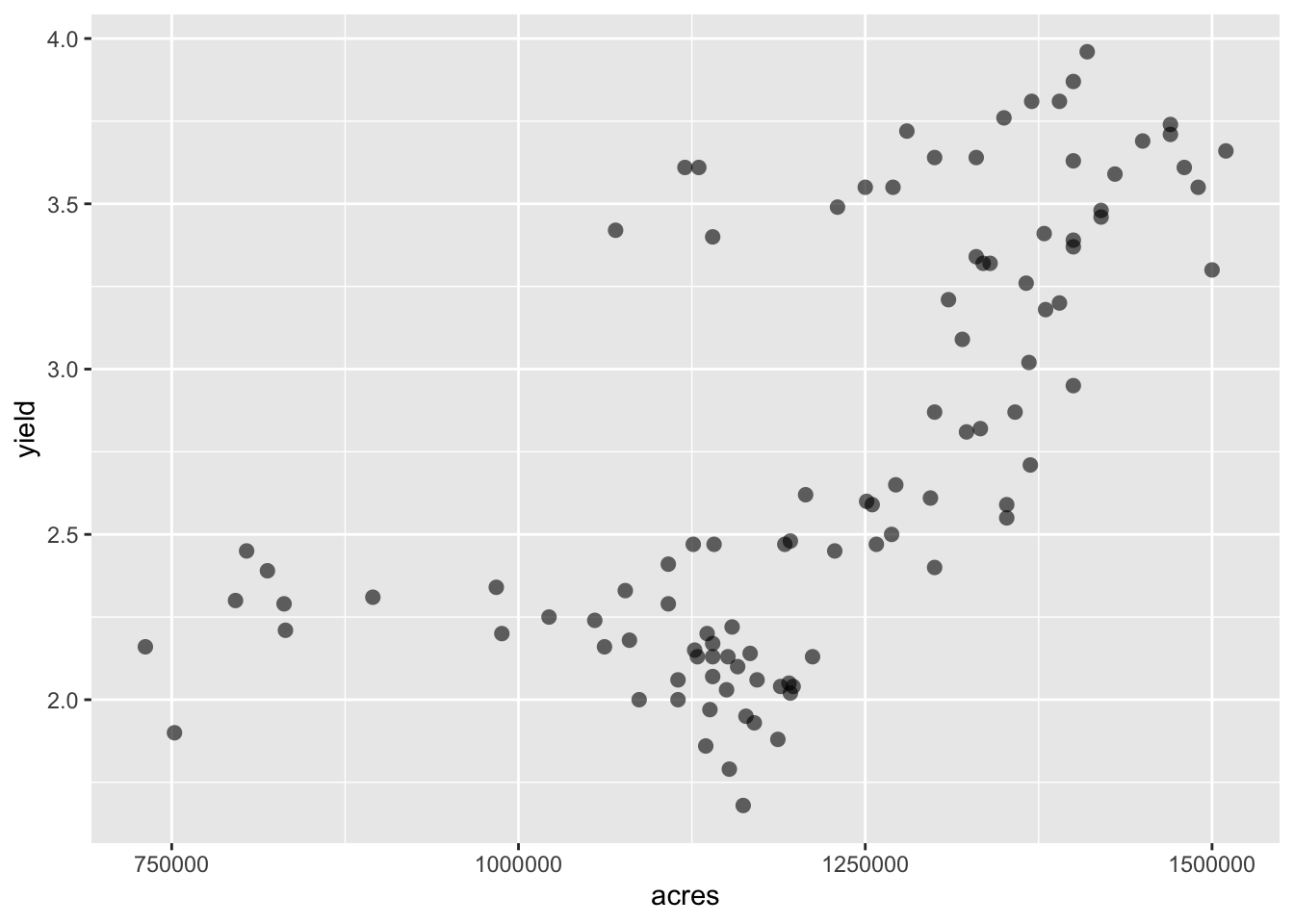
P <- ggplot(hay_idaho, aes(x = acres, y = yield)) +
geom_point(shape = 16, alpha = 0.6, size = 2.5)
P # Plots can be assigned to an object and then later added to
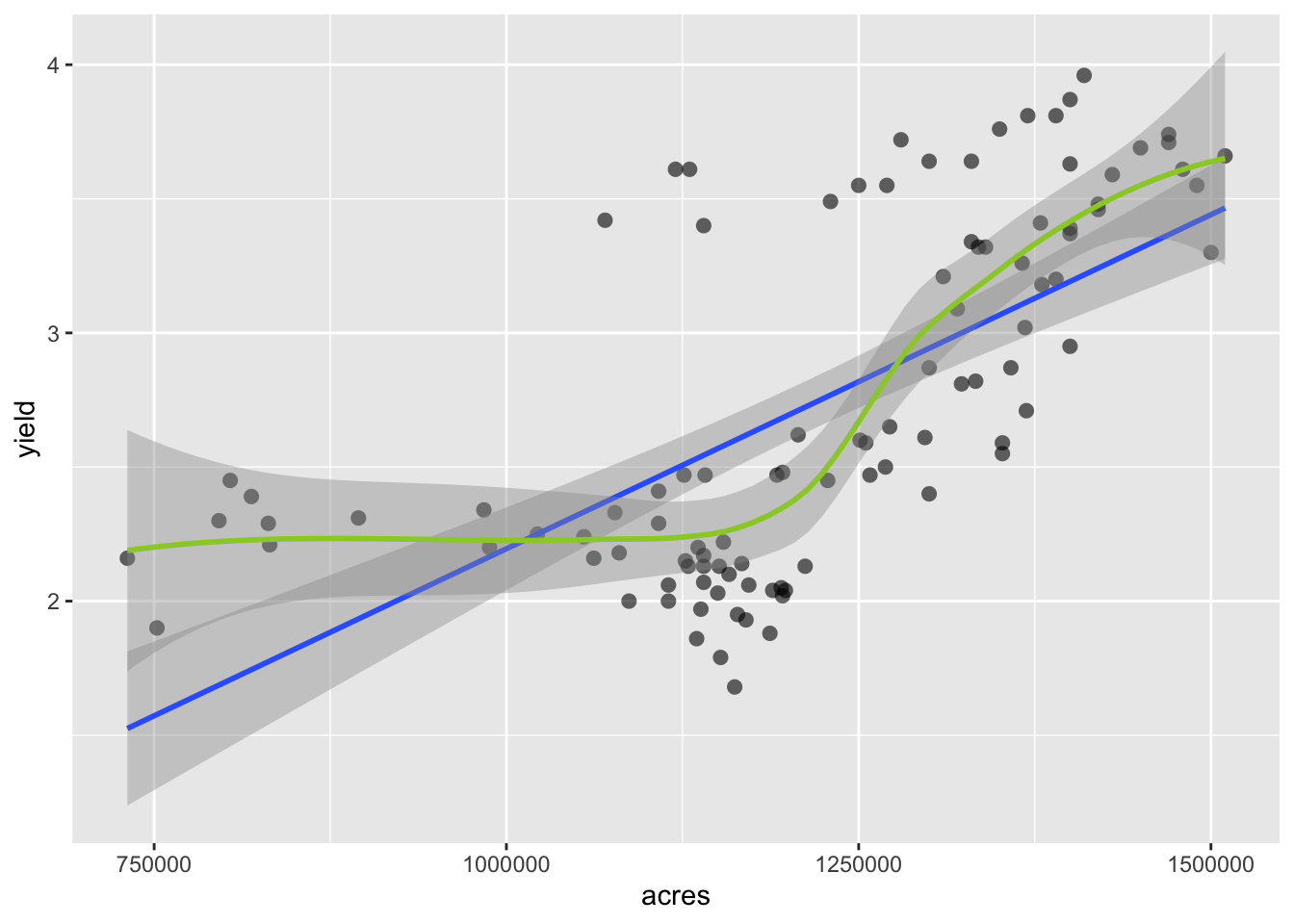
P +
geom_smooth(method = "lm") +
geom_smooth(method = "loess", col = "yellowgreen")
## `geom_smooth()` using formula 'y ~ x'
## `geom_smooth()` using formula 'y ~ x'
Adjusting non-data items
- Adjustments to the background, text size and other details are done in the
theme()argument command. - there are several pre-built themes:

Implementing a ggplot theme
facet.plot <- ggplot(hay_pnw) +
geom_line(aes(x = year, y = yield), col = "darkcyan") +
facet_wrap(. ~ state, nrow = 2, ncol = 2)
facet.plot + theme_minimal()
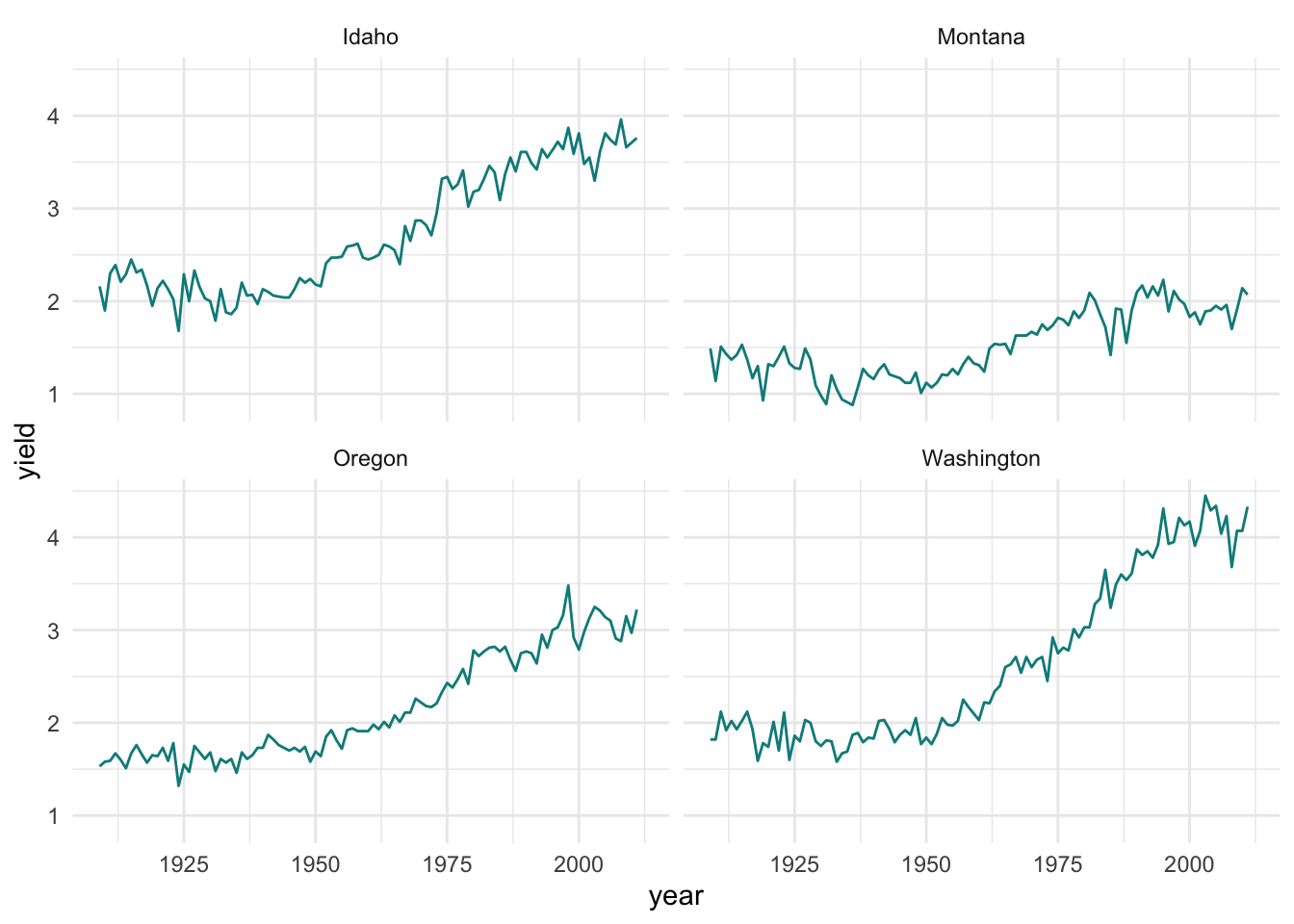
This is not exactly what I want. Larger labels on everything would be helpful, as well as an outline around each plot. And while we are at it, labeling the x-axis for each decade rather than every 25 years would be easier to interpret.
Adjusting text size
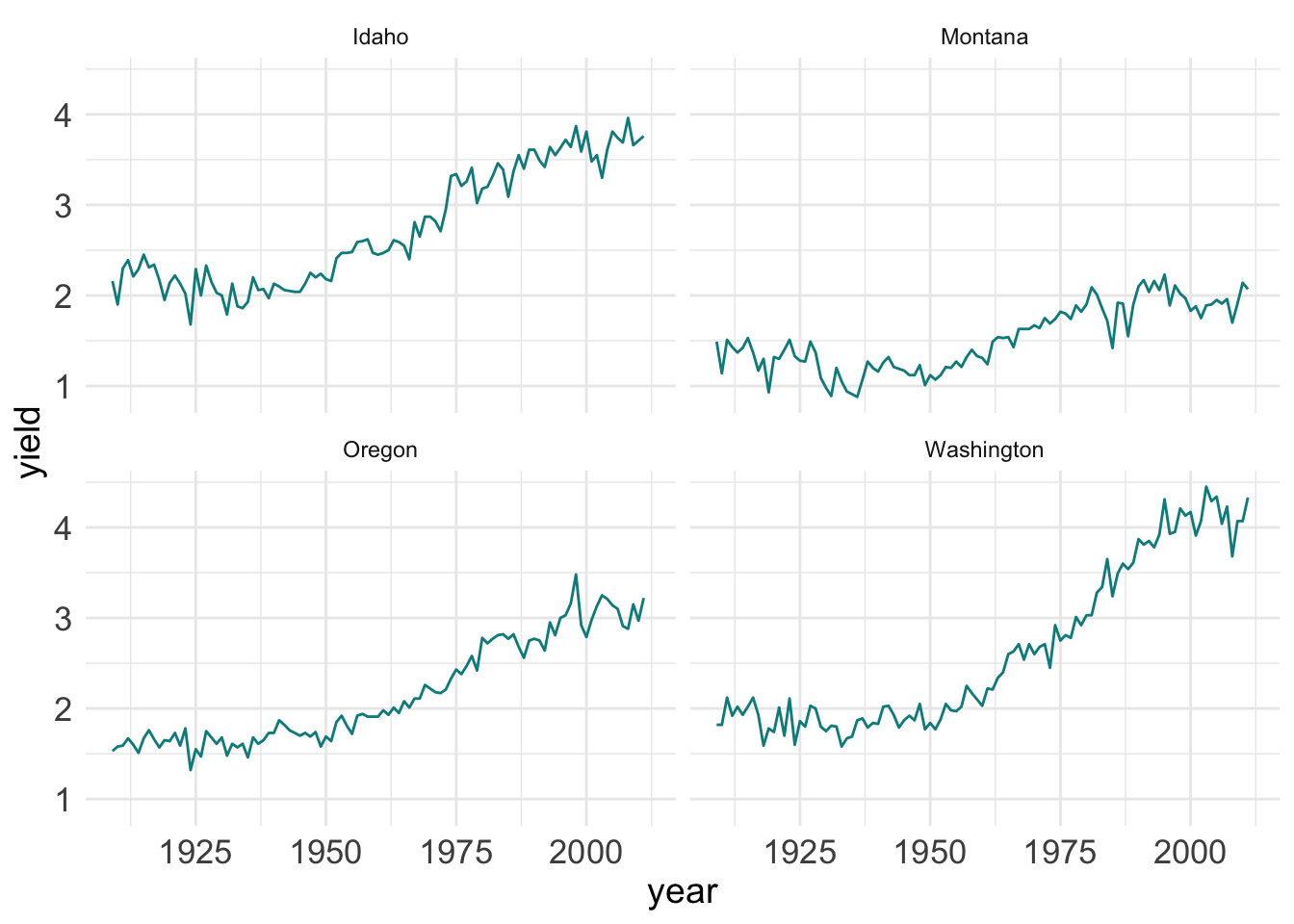
facet.plot +
theme_minimal() + # this must be added to plot before manual theme adjustments
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 13)
)
Adjusting the axis and gridlines
facet.plot +
scale_x_continuous(breaks = c(seq(1910, 2015, by = 10))) +
theme_minimal() +
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 13)
)
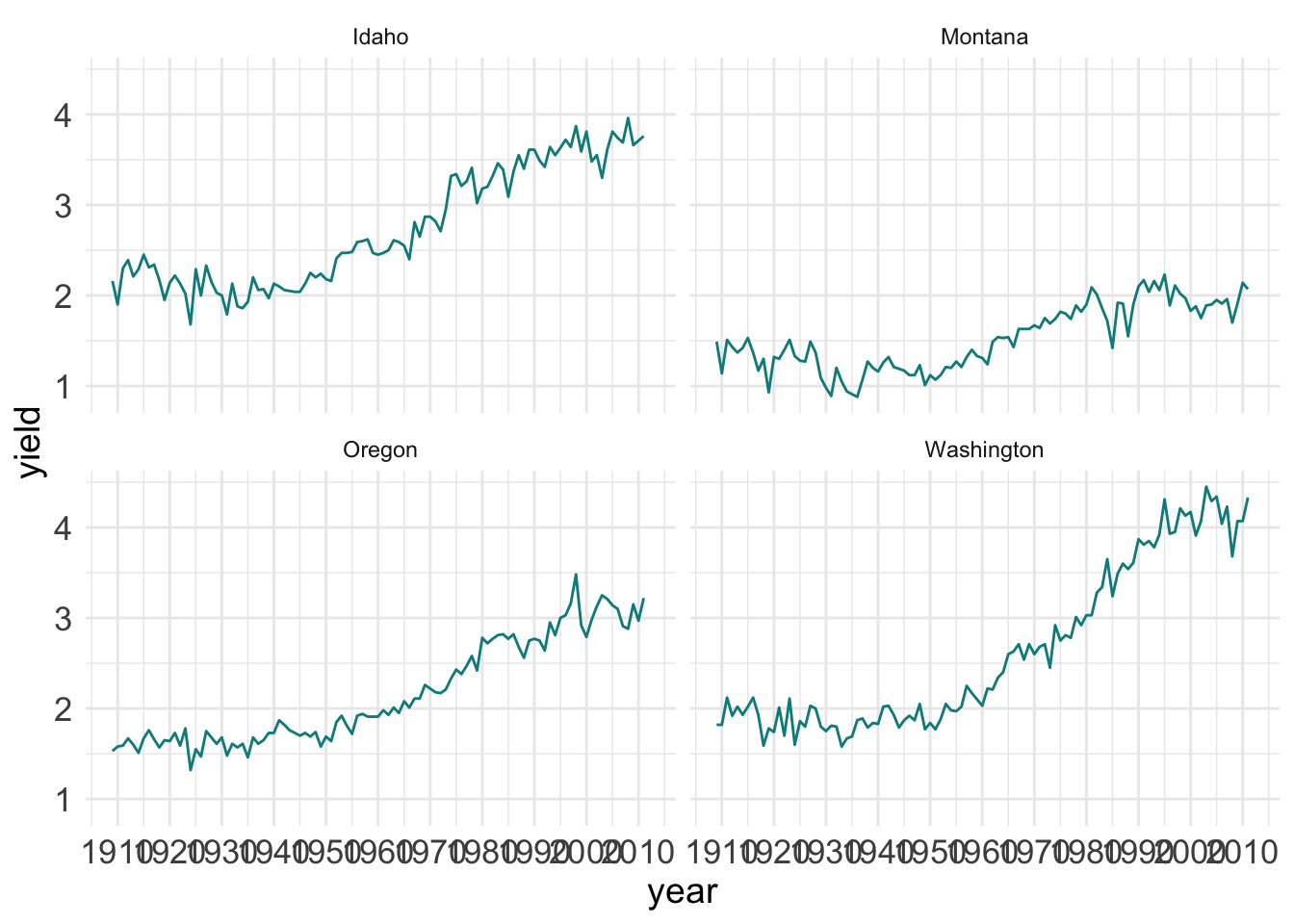
It looks like the years are spaced too close together and there are too many grid lines. This can all be fixed.
facet.plot +
scale_x_continuous(breaks = c(seq(1910, 2015, by = 10))) +
theme_minimal() + # this must be added to plot before manual theme adjustments
theme(
axis.title = element_text(size = 14), # adsjut title of the axes
axis.text = element_text(size = 13), # adjust axes tick labels
axis.text.x = element_text(angle = 45, hjust = 1), # adjust individual axis tick labels
panel.grid.minor = element_blank(), # remove minor grid lines (the major grid lines were retained)
panel.border = element_rect(colour = "black", fill = "transparent"), # create a border around each panel
strip.text = element_text(size = 15) # adjust panel labels (e.g. "Idaho")
)
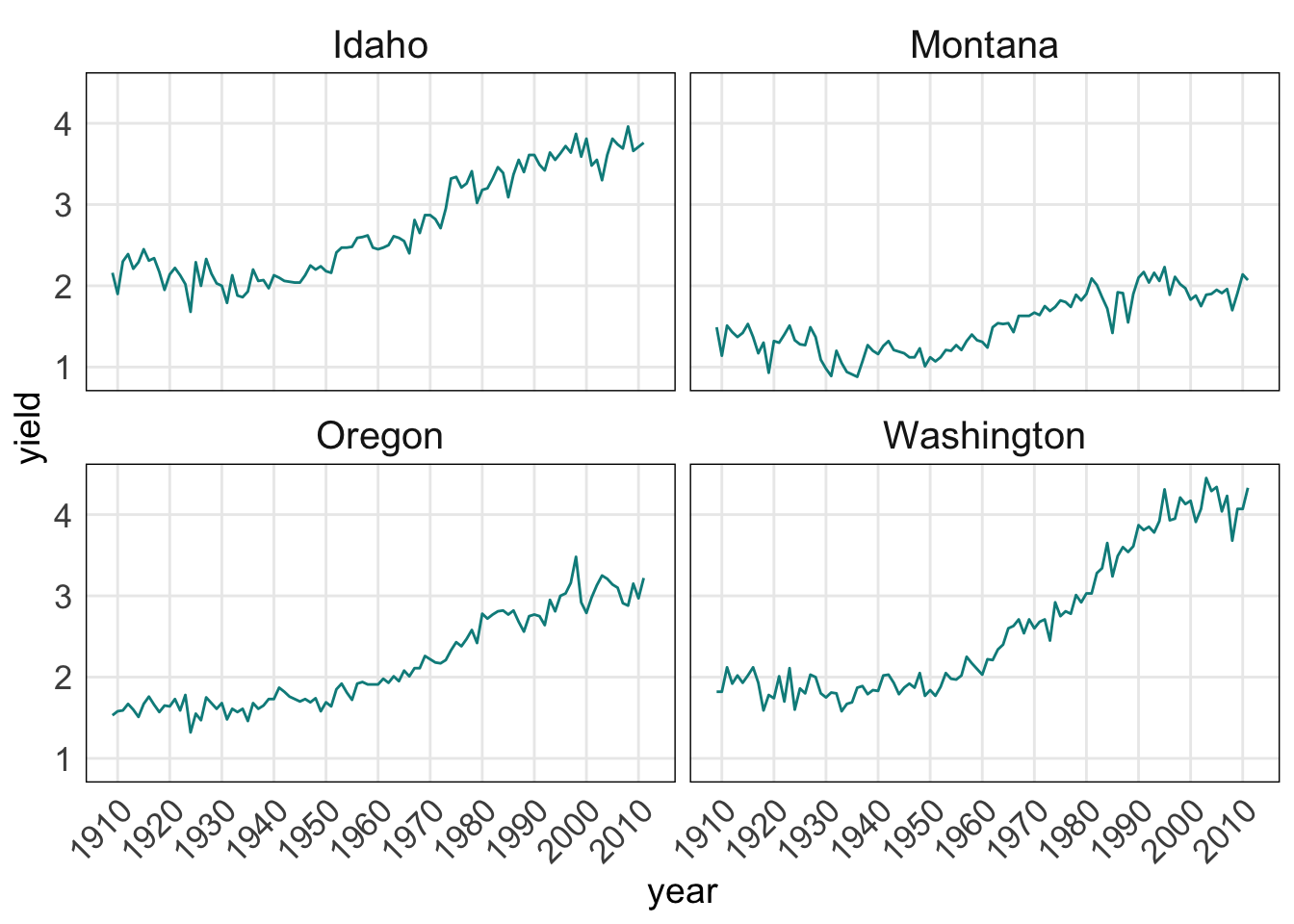
Adjusting the legend
ggplot(hay_pnw) +
geom_line(aes(x = year, y = yield, col = state)) +
scale_x_continuous(breaks = c(seq(1910, 2015, by = 10))) +
theme_bw()
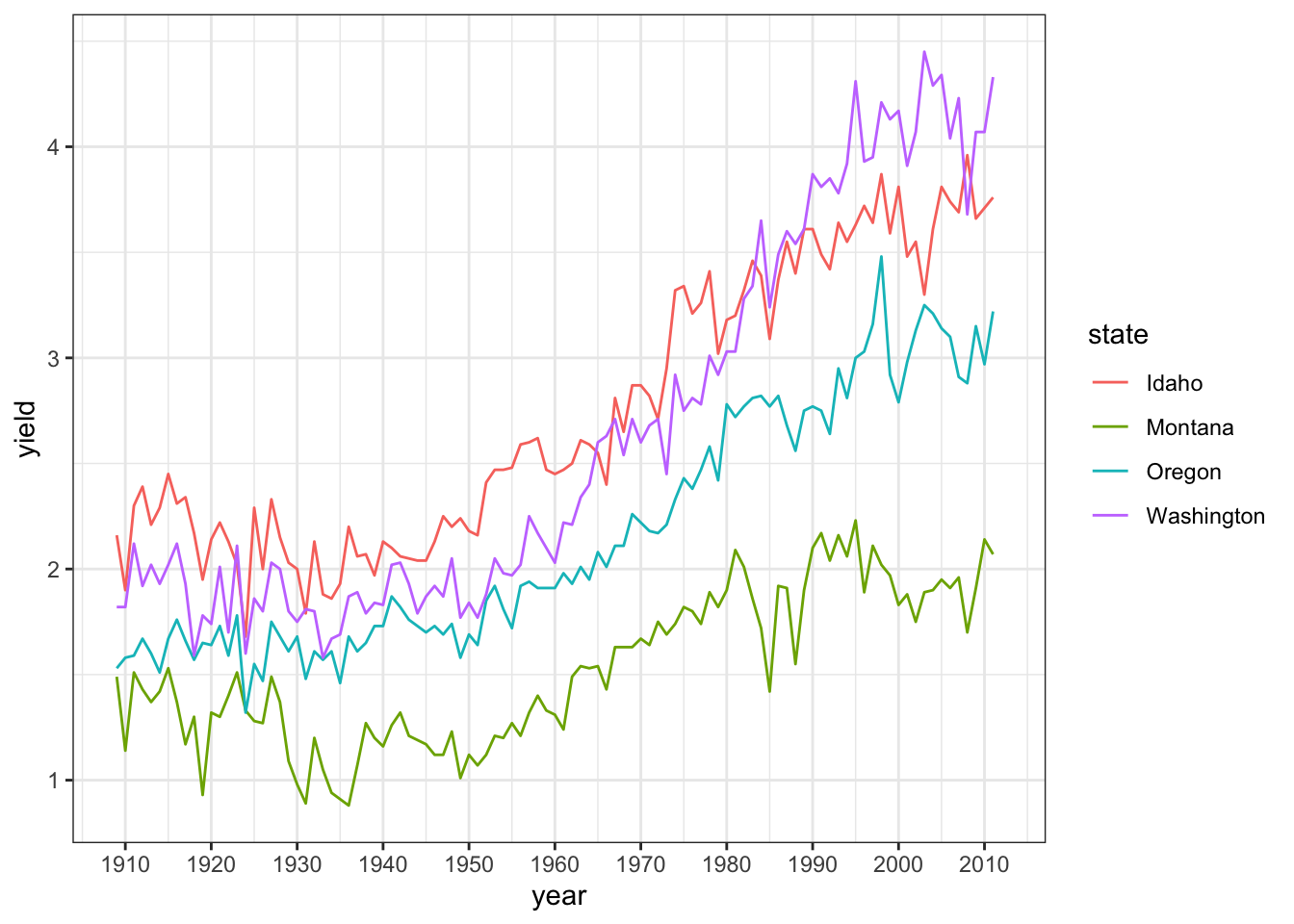
This looks okay, but it could be better. There is room to put the legend inside the plot and the text should probably be larger. Also, the legend title is not really needed.
ggplot(hay_pnw) +
geom_line(aes(x = year, y = yield, col = state)) +
scale_x_continuous(breaks = c(seq(1910, 2015, by = 10))) +
theme_bw() +
theme(
legend.position = c(0.15, 0.8), # give exact coordinates (ranging from 0 to 1)
legend.background = element_rect(colour = "black"),
legend.text = element_text(size = 12),
legend.title = element_blank()
)
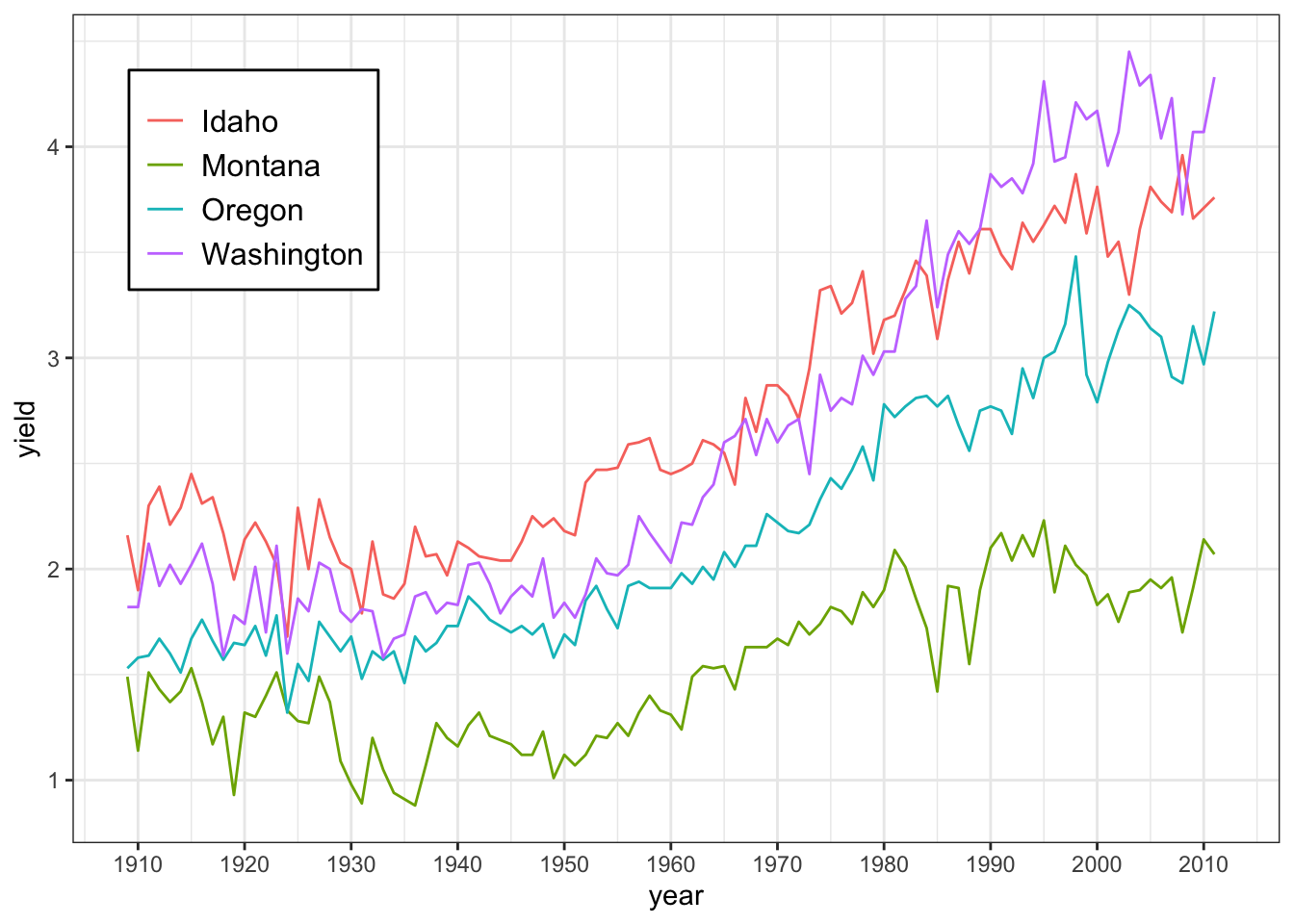
The legend can also be moved to locations outside the main plotting area with arguments such as legend.position = "top", legend.position = "left" and so on.
Summary
There are many options for adjusting text. You you will inevitable need to consult help files at some point.
A bit on colors
R has a rich set of colors and some excellent palettes that provide excellent contrast and/or work for color-blind individuals. Several are automatically loaded with ggplot2, such as the RColorBrewer:
library(RColorBrewer)
display.brewer.all()
![]()
The package viridis also has some nice color palettes that are widely used. Also check out the paletteer](https://emilhvitfeldt.github.io/paletteer/) and the accompanying R palette picker for a wider set of options.
Other Important Stuff
How to save plots
- The function
ggsave()is the recommended approach. - By default, it will save the the last plot created.
ggplot(hay_pnw) +
geom_line(aes(x = year, y = yield), col = "violetred") +
facet_wrap(. ~ state, nrow = 2, ncol = 2)
ggsave("PNW_hay_production.png", dpi = 300)
- other acceptable formats:
- .jpeg
- .tiff
- .bmp
- .svg
- .wmf (on windows machines)
- .eps
- .ps
- .tex
- .pd
- can specify the size of the saved plot (in centimeters, millimeters or inches) and the resolution in DPI (dots per square inch)
Other great geoms
| geom | purpose |
|---|---|
geom_violin() |
violin plot |
geom_ribbon() |
horizontal filled undulating band |
geom_bar() |
for creating bar plots |
geom_abline(), geom_hline(), geom_vline() |
for adding straight lines to a plot |
geom_map() |
part of the rich ecosystem of ggplot mapping functions |
geom_rug() |
adds rug plots to edges of a plot |
ggplot extensions
Many packages have been developed to extend ggplot’s functionality. Here are a few notable examples:
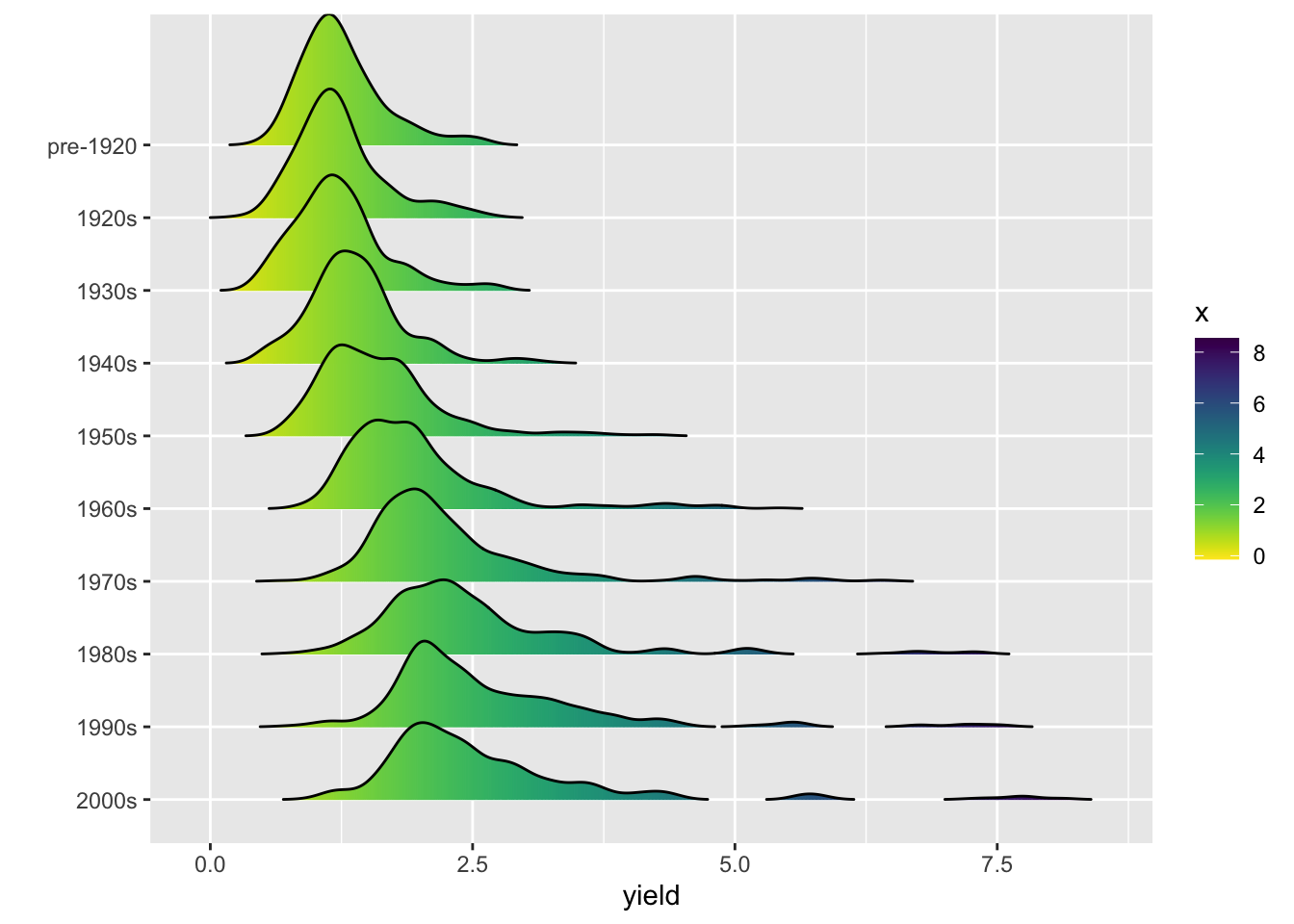
ggridge
# break hay data into decennial increments (roughly)
nass.hay$decade <- cut(nass.hay$year, breaks = c(1900, 1920, 1930, 1940, 1950, 1960, 1970, 1980, 1990, 2000, 2020), labels = c("pre-1920", "1920s", "1930s", "1940s", "1950s", "1960s", "1970s", "1980s", "1990s", "2000s"))
library(ggridges); library(viridis)
## Loading required package: viridisLite
ggplot(nass.hay, aes(x = yield, y = reorder(decade, desc(decade)), fill = ..x..)) +
geom_density_ridges_gradient(rel_min_height = 0.001) +
scale_fill_viridis(direction = -1) +
ylab("") # suppress the y-axis label
## Picking joint bandwidth of 0.129
plotly
library(plotly)
pl <- ggplot(nass.hay, aes(x = acres, y = yield, color = year)) +
geom_point() +
geom_point(aes(text = state)) +
#, shape = 21, col = "gray30", alpha = 0.6) +
#scale_fill_manual(values = alpha(rainbow(nlevels(nass.hay$state)), 0.5))
scale_color_gradientn(colors = alpha(rainbow(20), 0.5))
ggplotly(pl)
To save an interactive plot, use htmlwidgets::saveWidget().
gganimate example

Plot from “Learn gganimate”.
Other resources
vignette("ggplot2")
-
There are innumerable ggplot tutorials like this one on the interwebs, such as here, here, and here, among others
-
R Graphics Cookbook ($$). This book is written by Winston Chang, the main author of the ggplot2 package.
 * ggplot-focused data visualization workshop by Cédric Scherer
* ggplot-focused data visualization workshop by Cédric Scherer
ggplot can be fun
Plot of shots made by NBA Player Steph Curry, made using gganimate:
library(tidyr)
library(rjson)
# shot data for Stephen Curry
playerID <- 201939
shotURL <- paste("http://stats.nba.com/stats/shotchartdetail?CFID=33&CFPARAMS=2014-15&ContextFilter=&ContextMeasure=FGA&DateFrom=&DateTo=&GameID=&GameSegment=&LastNGames=0&LeagueID=00&Location=&MeasureType=Base&Month=0&OpponentTeamID=0&Outcome=&PaceAdjust=N&PerMode=PerGame&Period=0&PlayerID=",playerID,"&PlayerPosition=&PlusMinus=N&Position=&Rank=N&RookieYear=&Season=2014-15&SeasonSegment=&SeasonType=Regular+Season&TeamID=0&VsConference=&VsDivision=&mode=Advanced&showDetails=0&showShots=1&showZones=0", sep = "")
# import from JSON
shotData <- fromJSON(file = shotURL, method="C")
# unlist shot data, save into a data frame
shotDataf <- data.frame(matrix(unlist(shotData$resultSets[[1]][[3]]), ncol=24, byrow = TRUE))
# shot data headers
colnames(shotDataf) <- shotData$resultSets[[1]][[2]]
# covert x and y coordinates into numeric
shotDataf$LOC_X <- as.numeric(as.character(shotDataf$LOC_X))
shotDataf$LOC_Y <- as.numeric(as.character(shotDataf$LOC_Y))
shotDataf$SHOT_DISTANCE <- as.numeric(as.character(shotDataf$SHOT_DISTANCE))
# extract relevant data for plotting
shots_made <- shotDataf %>% filter(EVENT_TYPE == "Made Shot") %>% select(LOC_X, LOC_Y) %>%
drop_na()
# set colors
cols <- rep(c("blue", "blueviolet","cyan", "magenta"), ceiling(653/4))[1:653]
# make plot!
ggplot(shots_made, aes(x=LOC_X, y=LOC_Y)) +
geom_segment(aes(x = LOC_X, y = LOC_Y, xend = 0, yend = 0),
col = cols) +
geom_point(shape = 16, col = "white", alpha = 0.8, size = 2) +
ylim(c(-10, 300)) +
theme_classic() +
theme(panel.background = element_rect(fill = "black"),
plot.background = element_rect(fill = "black"),
plot.title = element_text(colour = "white", size = 20, hjust = 0.5, face = "bold"),
axis.text = element_blank()
)
