Introduction to Data Cleaning with the Tidyverse

knitr::opts_chunk$set(echo = TRUE, eval=TRUE, include=TRUE)
If your file management system looks like this (no guilt, we have all been there), this is suggestive of some rather ad hoc approaches to data cleaning. As you work with your data, you will discover errors (this is a universal experience). How do you fix those?
Newer scientists are likely to use point-and-click software like spreadsheet. The problem with this approach is that there is no record of your exact action, so you save a new copy of the data set with a new name. And on it goes. There are other ways to fix data errors using a transparent process that can avoid these multiplying data sets, or clarify what distinguishes one data set from another.
Introduction
Equip everyone regardless of means, to participate in the global economy that rewards data literacy.
The Tidyverse is a collection of packages designed to help data users work more efficiently with their data. At the time of writing this, the Tidyverse consists of over 25 packages with diverse that range from connecting to google sheets to working with databases.
Most Tidyverse functions have equivalent functions in base R (that is, the set of function that load automaticallty when R is launched). The difference is that tidyverse functions are often faster, the syntax may be clearer and easier to use (no guarantee, tho), and most importantly, they all follow a similar structure and the results are consistent across packages (within limits, this depends on the nature of the function).
Technical note: the Tidyverse uses “non-standard evaluation” (NSE) principles in its functions. NSE is a complicated topic, but in brief, it means that tidyverse function follow different rules than base R function. Tidyverse functions are easier to use, but, if you are used to base R functions, it will take some effort to reorient your R programming brain. Just a friendly warning before we dive in!
Tidyverse Packages
Abbreviated list of packages useful for data cleaning
As mentioned, the Tidyverse is a collection of many packages. Below is a table of many very useful packages for data cleaning. This tutorial does not address all these packages, but they are worth checking out, depending on your data analytic needs.
| Package | Purpose |
|---|---|
| dplyr | main package for organizing data |
| forcats | for managing categorical data |
| janitor | also not part of the tidyverse, has useful data cleaning functions |
| lubridate | for manipulating dates |
| purrr | advanced broadcasting of functions across data |
| readr | for reading in tabular data |
| readxl | for reading in MS excel sheets |
| stringr | for managing “strings” (character variables) |
| tidyr | to reshape data and handle missing values |
 Honorable mention: data.table, which provides advanced handling functions for data.frames. While not part of the tidyverse officially, it is very efficient.
Honorable mention: data.table, which provides advanced handling functions for data.frames. While not part of the tidyverse officially, it is very efficient.
Packages Used in This Tutorial
library(readr)
library(data.table)
library(dplyr)
##
## Attaching package: 'dplyr'
## The following objects are masked from 'package:data.table':
##
## between, first, last
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
library(tidyr)
The warning messages indicate when there are identical function names across different packages. The last package loaded will override functions from previous-loaded packages when such conflicts arise. So, the order of how packages are loaded can matter. However, any function can be accessed by add a prefix of the package name and two colons to a particular function:
base::intersect(...)
Load Data
read_csv() v read.csv()
How do the functions read.csv() (from base R) and read_csv (from readr) differ in how they read in data?
For this tutorial, 3 data sets will be used, “nass_cotton”, “nass_barley”, and “nass_barley”. These are historical data sets from the National Agricultural Statistics Service that indicate the total acreage and yield for the crops indicated for each each U.S. state and year from 1866 to 2011. They can be accessed with the package ‘agridat’.
These data sets were originally obtained as thus from Agridat and saved to separate files for this tutorial.
library(agridat) # a package of agricultural data sets
data("nass.corn")
data("nass.barley")
data("nass.cotton")
write.csv(nass.corn, "nass_corn.csv", row.names = F)
write.csv(nass.barley, "nass_barley.csv", row.names = F)
write.csv(nass.cotton, "nass_cotton.csv", row.names = F)
When using the RStudio’s point-and-click interface to load tabular data, it will typically call the function read_csv() for CSV files and read_delim() for text files. read_csv() is a wrapper for read_delim() with different default arguments.
These arguments can be called using the command line interface as well.
nass_barley <- read.csv("nass_barley.csv")
nass_cotton <- read_csv("nass_cotton.csv")
## Rows: 2338 Columns: 4
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (1): state
## dbl (3): year, acres, yield
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
# contrast functions read.csv and read_csv
str(nass_barley)
## 'data.frame': 4839 obs. of 4 variables:
## $ year : int 1866 1866 1866 1866 1866 1866 1866 1866 1866 1866 ...
## $ state: chr "Connecticut" "Illinois" "Indiana" "Iowa" ...
## $ acres: num 1000 96000 11000 66000 2000 10000 34000 7000 21000 20000 ...
## $ yield: num 22.5 23.4 23 22 23 23.5 21.5 25.5 26 26 ...
str(nass_cotton)
## spec_tbl_df [2,338 × 4] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## $ year : num [1:2338] 1866 1866 1866 1866 1866 ...
## $ state: chr [1:2338] "Alabama" "Arkansas" "Florida" "Georgia" ...
## $ acres: num [1:2338] 977000 489000 155000 895000 1020000 ...
## $ yield: num [1:2338] 120 198 123 122 57 86 124 116 102 121 ...
## - attr(*, "spec")=
## .. cols(
## .. year = col_double(),
## .. state = col_character(),
## .. acres = col_double(),
## .. yield = col_double()
## .. )
## - attr(*, "problems")=<externalptr>
A Bit on the Tibble
A tibble is an object type created for the Tidyverse. This is the object type imported by ‘readr’ and returned by many tidyverse functions. The tibble is like a data frame with a few other rules. Tibbles have a simplified printing function if you accidentally type in the object name - it will only print the first 10 lines and will truncate the number of columns to fit your screen. If you type the object name of a traditional data frame, the first 1000 lines of data are printed!
To create a new tibble, you can use the function tribble() to create a tibble by rows, or the function tibble() for building a tibble column-wise, or as.tibble() to coerce an object to a tibble.

There are a few aspects of tibbles that are fundamentally different from a standard data frame. Tibbles do not handle row names well and many tidyverse functions discard that information. This is often just fine - row names are computationally expensive to set and maintain. Also, it can be difficult to filter and sort data based on row names. However, many other (non-tidyverse) package functions rely on the rownames attribute, so be aware of this behavior when using tidyverse functions on data sets.
Function details
-
the most noticeable difference between
read.csv()andread_csv()is thatread.csv()’s default behavior is to replace all spaces and special characters in column headers with a “.”, whileread_csv()keeps all that information. Additionally,read_csv()tends to handle dates a bit better thanread.csv(). -
read_delim()and its wrappers are faster than base versions (e.g.read.csv()), however, the difference will not be noticeable for small files (< 10 Mb) -
read_delim()offers additional flexibility such removing trailing and leading white space (" hello" and “hello “) and automatically deleting empty rows (these things occur by default). -
specific values for missing data that are not “NA” (e.g. “-9”, “.") can be set with with argument
na = ... -
the function can be sped up by setting values for the
col_types = ...argument (e.g. character, numeric, date, et cetera):
nass_corn <- read_csv("nass_corn.csv", col_types = cols(col_integer(), col_character(), col_double(), col_double()))
str(nass_corn)
## spec_tbl_df [6,381 × 4] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## $ year : int [1:6381] 1866 1866 1866 1866 1866 1866 1866 1866 1866 1866 ...
## $ state: chr [1:6381] "Alabama" "Arkansas" "California" "Connecticut" ...
## $ acres: num [1:6381] 1050000 280000 42000 57000 200000 ...
## $ yield: num [1:6381] 9 18 28 34 23 9 6 29 36.5 32 ...
## - attr(*, "spec")=
## .. cols(
## .. year = col_integer(),
## .. state = col_character(),
## .. acres = col_double(),
## .. yield = col_double()
## .. )
## - attr(*, "problems")=<externalptr>
Note: if you have truly huge data, consider the fread() function in ‘data.table’, storing the data in database and using ‘dbplyr’ to access it, or using the ‘arrow’ package.
Data selection and subsetting
Using ‘dplyr’
Column selection
The select() function is used for column selection and has the general structure: select(data, variables_to_select).
Select only the columns that you want
barley_acres <- select(nass_barley, state, year, acres)
str(barley_acres)
## 'data.frame': 4839 obs. of 3 variables:
## $ state: chr "Connecticut" "Illinois" "Indiana" "Iowa" ...
## $ year : int 1866 1866 1866 1866 1866 1866 1866 1866 1866 1866 ...
## $ acres: num 1000 96000 11000 66000 2000 10000 34000 7000 21000 20000 ...
head(barley_acres)
## state year acres
## 1 Connecticut 1866 1000
## 2 Illinois 1866 96000
## 3 Indiana 1866 11000
## 4 Iowa 1866 66000
## 5 Kansas 1866 2000
## 6 Kentucky 1866 10000
Select the columns you do not want:
barley_yield <- select(nass_barley, -acres)
str(barley_yield)
## 'data.frame': 4839 obs. of 3 variables:
## $ year : int 1866 1866 1866 1866 1866 1866 1866 1866 1866 1866 ...
## $ state: chr "Connecticut" "Illinois" "Indiana" "Iowa" ...
## $ yield: num 22.5 23.4 23 22 23 23.5 21.5 25.5 26 26 ...
barley_yield2 <- select(nass_barley, -(c(acres, yield))) # multiple columns
str(barley_yield2)
## 'data.frame': 4839 obs. of 2 variables:
## $ year : int 1866 1866 1866 1866 1866 1866 1866 1866 1866 1866 ...
## $ state: chr "Connecticut" "Illinois" "Indiana" "Iowa" ...
Sorting data
Sort functions follow this structure: sort(data, variables_to_sort)
Sort across one column:*
corn_year <- arrange(nass_corn, state)
head(corn_year)
## # A tibble: 6 × 4
## year state acres yield
## <int> <chr> <dbl> <dbl>
## 1 1866 Alabama 1050000 9
## 2 1867 Alabama 1400000 13
## 3 1868 Alabama 1420000 12.5
## 4 1869 Alabama 1430000 12
## 5 1870 Alabama 1330000 13.5
## 6 1871 Alabama 1480000 11
Sort across several columns
corn_year_yield <- arrange(nass_corn, year, yield)
head(corn_year_yield)
## # A tibble: 6 × 4
## year state acres yield
## <int> <chr> <dbl> <dbl>
## 1 1866 Georgia 1770000 6
## 2 1866 South Carolina 950000 6.5
## 3 1866 Alabama 1050000 9
## 4 1866 Florida 125000 9
## 5 1866 Mississippi 800000 9.5
## 6 1866 North Carolina 1675000 9.5
Sort in reverse order
corn_year_yield_rev <- arrange(nass_corn, desc(year), desc(yield))
head(corn_year_yield_rev)
## # A tibble: 6 × 4
## year state acres yield
## <int> <chr> <dbl> <dbl>
## 1 2011 Washington 125000 225
## 2 2011 Oregon 51000 215
## 3 2011 California 150000 185
## 4 2011 Idaho 120000 185
## 5 2011 Arizona 32000 180
## 6 2011 New Mexico 43000 180
Row selection
Based on their position or index (where the first row = 1):
corn_first100 <- slice(nass_corn, 100) # grab first 100 rows
Filtering Data
(actually a type of row selection)
Filtering enables users to select all columns of data based on a condition within a row or rows. The filter() function has the formulation: select(data, condition_for_filter).
Filter based on one variable and exact match
corn_AZ <- filter(nass_corn, state == "Arizona")
head(corn_AZ)
## # A tibble: 6 × 4
## year state acres yield
## <int> <chr> <dbl> <dbl>
## 1 1882 Arizona 3000 21
## 2 1883 Arizona 3000 20
## 3 1884 Arizona 3000 21
## 4 1885 Arizona 3000 22
## 5 1886 Arizona 3000 22
## 6 1887 Arizona 3000 19
Filter for matching several items:
barley_pnw <- filter(nass_barley, state %in% c("Idaho", "Oregon", "Washington"))
head(barley_pnw)
## year state acres yield
## 1 1869 Oregon 6000 35.0
## 2 1870 Oregon 7000 30.5
## 3 1871 Oregon 8000 28.5
## 4 1872 Oregon 11000 30.5
## 5 1873 Oregon 15000 33.0
## 6 1874 Oregon 17000 29.0
Filter by numerical cut-off:
(first, look at summary of data)
summary(nass_cotton$acres)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 100 118000 550000 1206414 1431500 17749000
nass_cotton_high <- filter(nass_cotton, acres < 550000)
Logical operators in R:
| symbol | meaning |
|---|---|
| < | less than |
| <= | less than or equal to |
| > | greater than |
| >= | greater than or equal to |
| == | exactly equal to |
| != | not equal to |
| !x | Not x |
| x | y |
| x & y | x AND y |
Combine multiple filters
(and plot the output)
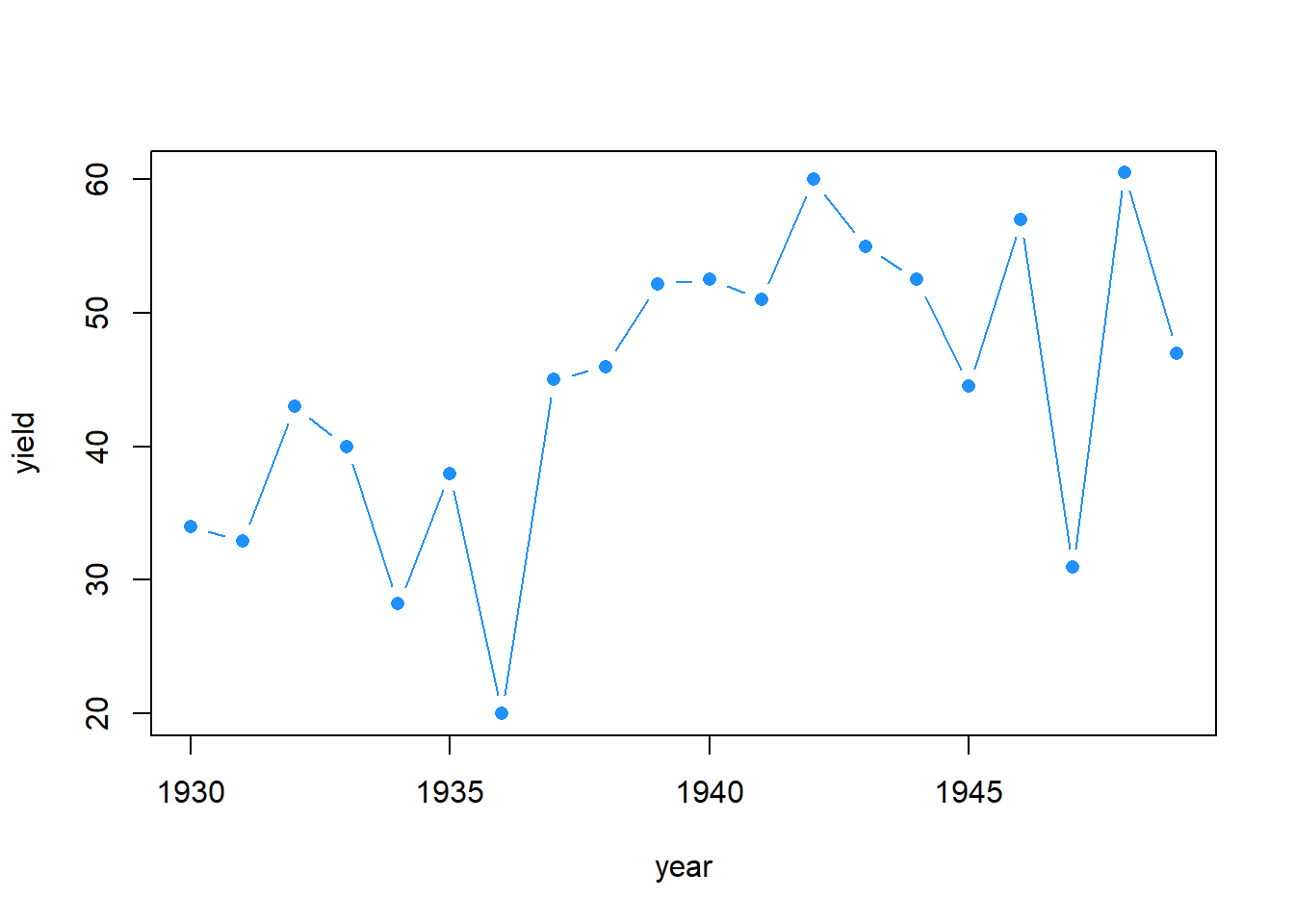
corn_Iowa_prewar <- filter(nass_corn, year < 1950 & year >= 1930 & state == "Iowa")
with(corn_Iowa_prewar, plot(year, yield, col = "dodgerblue", pch = 16, type = "b"))
Data transformations
The mutate() function to define new variables
Mutate functions have the formulation: mutate(data, new_variable_name = data_transformation). New variables are declared on the left and the right provides the operations for variable creation itself. Many operations are allowed within a mutate function:
barley_HI <- mutate(nass_barley,
harvest_index = acres/yield,
Year = as.factor(year),
Is_high = acres > median(acres),
crop = "barley",
relative_yld = yield/mean(yield))
head(barley_HI)
## year state acres yield harvest_index Year Is_high crop relative_yld
## 1 1866 Connecticut 1000 22.5 44.44444 1866 FALSE barley 0.6494957
## 2 1866 Illinois 96000 23.4 4102.56410 1866 TRUE barley 0.6754755
## 3 1866 Indiana 11000 23.0 478.26087 1866 FALSE barley 0.6639290
## 4 1866 Iowa 66000 22.0 3000.00000 1866 TRUE barley 0.6350625
## 5 1866 Kansas 2000 23.0 86.95652 1866 FALSE barley 0.6639290
## 6 1866 Kentucky 10000 23.5 425.53191 1866 FALSE barley 0.6783622
Renaming variables
The rename() function renames columns using this notation: rename(dataframe, newname = "oldname"):
barley_HI <- rename(barley_HI, high_acres = "Is_high")
str(barley_HI)
## 'data.frame': 4839 obs. of 9 variables:
## $ year : int 1866 1866 1866 1866 1866 1866 1866 1866 1866 1866 ...
## $ state : chr "Connecticut" "Illinois" "Indiana" "Iowa" ...
## $ acres : num 1000 96000 11000 66000 2000 10000 34000 7000 21000 20000 ...
## $ yield : num 22.5 23.4 23 22 23 23.5 21.5 25.5 26 26 ...
## $ harvest_index: num 44.4 4102.6 478.3 3000 87 ...
## $ Year : Factor w/ 146 levels "1866","1867",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ high_acres : logi FALSE TRUE FALSE TRUE FALSE FALSE ...
## $ crop : chr "barley" "barley" "barley" "barley" ...
## $ relative_yld : num 0.649 0.675 0.664 0.635 0.664 ...
Splitting and Merging Columns
Combine multiple columns into one with the function unite(). Column data are pasted together as text, not added as numbers. The columns used to make the new variables are automatically discarded (this behavior can be stopped with the argument remove = F).
unite() functions have this formulation: unite(data, new_var_name, vars_to_select).
cotton_nass_new <- unite(nass_cotton, "ID", state, year)
head(cotton_nass_new)
## # A tibble: 6 × 3
## ID acres yield
## <chr> <dbl> <dbl>
## 1 Alabama_1866 977000 120
## 2 Arkansas_1866 489000 198
## 3 Florida_1866 155000 123
## 4 Georgia_1866 895000 122
## 5 Louisiana_1866 1020000 57
## 6 Mississippi_1866 1668000 86
The Opposite function,separate(), has this structure: separate(data, col_to_split, names_of_new_cols)
cotton_nass_new2 <- separate(cotton_nass_new,
col = ID,
into = c("STATE", "YEAR"),
sep = "_",
remove = F)
head(cotton_nass_new2)
## # A tibble: 6 × 5
## ID STATE YEAR acres yield
## <chr> <chr> <chr> <dbl> <dbl>
## 1 Alabama_1866 Alabama 1866 977000 120
## 2 Arkansas_1866 Arkansas 1866 489000 198
## 3 Florida_1866 Florida 1866 155000 123
## 4 Georgia_1866 Georgia 1866 895000 122
## 5 Louisiana_1866 Louisiana 1866 1020000 57
## 6 Mississippi_1866 Mississippi 1866 1668000 86
These last two functions are often quite useful - for pulling apart complicated ID columns, or for creating a unique identifier that can later be used for merging two data sets together.
Merging data sets with ‘dplyr’
Merging different data sets by a common variable is a major advantage to using automated software. Doing this manually in spreadsheet programs is nearly impossible to do without introducing errors. There are 6 different merging or joining functions which differ in what data they keep and what are discarded.
Standard join functions
For merging 2 data frames. Join functions are first and foremost defined by the rows they return. The function use a common ID, which can be automatically detected or explicitly declared, to match observations in A with observations in B and returned a merged data frame. In most cases, all columns from both data frames are returned.
The general formulation for join functions: type_join(x_data, y_data, ID_key_variable)

The column(s) used for ID are only included once in the merged data frame, that is, the duplicated key column is dropped. However, if there are other (non-ID) columns with duplicated names across the data sets, the duplicate columns from first data set listed are given a “.x” suffix and the duplicate columns from second data set listed are given a “.y” suffix.
1. The full_join():
-
always the safest option for avoiding data removal (everything is returned)
-
in this example, the full join will produce matches when both the year and state of a single observation match the year and state, and non-matches are appended to the resulting object.
-
Note: for the rest of the joins, which employ a filtering step, I recommend you always use a single unique identifier (called a “key”) for the joins. You can use multiple keys, but a single unique key is the clearest way to join data without generating duplicate observations.
-
When there is no match, the rows from each data set are still returned and filled with NA for columns from the other data set.
# first create a unique identifier variable
cotton_nass_new <- unite(nass_cotton, "ID", state, year)
barley_nass_new <- unite(nass_barley, "ID", state, year)
#equivalent models - this is only true for full_joins
cotton_barley_all <- full_join(nass_cotton, nass_barley, by = c("year", "state"))
cotton_barley_all2 <- full_join(cotton_nass_new, barley_nass_new, by = "ID")
# compare dimensions after the joins - what is kept?
dim(cotton_barley_all)
## [1] 5935 6
dim(nass_cotton)
## [1] 2338 4
dim(nass_barley)
## [1] 4839 4
2. The inner_join():
This function only keeps what is common between data sets:
cotton_barley_inner <- inner_join(cotton_nass_new, barley_nass_new, by = "ID")
dim(cotton_barley_inner)
## [1] 1242 5
head(cotton_barley_inner)
## # A tibble: 6 × 5
## ID acres.x yield.x acres.y yield.y
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Tennessee_1866 649000 121 4000 23
## 2 Texas_1866 490000 325 3000 16
## 3 Tennessee_1867 611000 137 4000 19
## 4 Texas_1867 491000 195 2000 20
## 5 Tennessee_1868 520000 170 4000 18
## 6 Texas_1868 581000 197 2000 22
- several column names duplicates resulted in the addition of ".x" to duplicate columns from the cotton data set and ".y" to duplicate columns from the barley data set.
colnames(cotton_barley_inner)[2:5] <- c("cotton_acres", "cotton_yield", "barley_acres", "barley_yield")
3. The left_join():
This function matches all rows for observations found in the tibble listed on the left:
cotton_barley_left <- left_join(cotton_nass_new, barley_nass_new, by = "ID")
dim(cotton_barley_left)
## [1] 2338 5
4. The right_join():
This function matches all rows for observations found in the tibble listed on the left:
cotton_barley_right <- right_join(cotton_nass_new, barley_nass_new, by = "ID")
dim(cotton_barley_right)
## [1] 4839 5
5. The semi_join():
This function is like inner_join() (only rows with keys in both data sets are kept) except it does not keep any data from the data set listed second:
cotton_barley_semi <- semi_join(cotton_nass_new, barley_nass_new, by = "ID")
dim(cotton_barley_semi)
## [1] 1242 3
str(cotton_barley_semi)
## tibble [1,242 × 3] (S3: tbl_df/tbl/data.frame)
## $ ID : chr [1:1242] "Tennessee_1866" "Texas_1866" "Tennessee_1867" "Texas_1867" ...
## $ acres: num [1:1242] 649000 490000 611000 491000 520000 581000 502000 720000 536000 922000 ...
## $ yield: num [1:1242] 121 325 137 195 170 197 145 195 203 255 ...
str(cotton_barley_inner)
## tibble [1,242 × 5] (S3: tbl_df/tbl/data.frame)
## $ ID : chr [1:1242] "Tennessee_1866" "Texas_1866" "Tennessee_1867" "Texas_1867" ...
## $ cotton_acres: num [1:1242] 649000 490000 611000 491000 520000 581000 502000 720000 536000 922000 ...
## $ cotton_yield: num [1:1242] 121 325 137 195 170 197 145 195 203 255 ...
## $ barley_acres: num [1:1242] 4000 3000 4000 2000 4000 2000 4000 2000 4000 2000 ...
## $ barley_yield: num [1:1242] 23 16 19 20 18 22 19 22 18.5 20.5 ...
6. The anti_join():
Observations from first data set that do not match the second data set are kept:
cotton_barley_anti <- anti_join(cotton_nass_new, barley_nass_new, by = "ID")
dim(cotton_barley_anti)
## [1] 1096 3
Question: What happens when there are multiple matches for an identifier when using full_join(), left_join() or right_join()?
Answer: Each time there is a new match between the keys of two data frames, a new row is added for the matched data - effectively all pairwise matches are added to the data set. There may be circumstances when this is advantageous, but usually, it is an unexpected surprise when a full_join() results in new data set several magnitudes larger than the two data sets combined.
Set operations
These are different from standard joins. They compare across vectors (not data.frames/tibbles) and return what has been specified in the statement. Duplicates are always discarded. The purpose of set operations is to determine common or uncommon observations, not merge two different data sets.
Common set operations

# states with barley data but no cotton data
setdiff(nass_barley$state, nass_cotton$test)
## Warning: Unknown or uninitialised column: `test`.
## [1] "Connecticut" "Illinois" "Indiana" "Iowa"
## [5] "Kansas" "Kentucky" "Maine" "Massachusetts"
## [9] "Michigan" "Minnesota" "Missouri" "Nebraska"
## [13] "New Hampshire" "New York" "Ohio" "Pennsylvania"
## [17] "Rhode Island" "Tennessee" "Texas" "Vermont"
## [21] "Wisconsin" "California" "Oregon" "Nevada"
## [25] "Colorado" "Virginia" "Arizona" "Idaho"
## [29] "Montana" "New Mexico" "North Dakota" "South Dakota"
## [33] "Utah" "Washington" "Maryland" "Oklahoma"
## [37] "Wyoming" "New Jersey" "North Carolina" "South Carolina"
## [41] "West Virginia" "Arkansas" "Delaware" "Georgia"
## [45] "Mississippi" "Alabama" "Alaska"
# states with cotton data, but no barley data
setdiff(nass_cotton$state, nass_barley$state)
## [1] "Florida" "Louisiana" "Otherstates"
# states that grow both
intersect(nass_cotton$state, nass_barley$state)
## [1] "Alabama" "Arkansas" "Georgia" "Mississippi"
## [5] "North Carolina" "South Carolina" "Tennessee" "Texas"
## [9] "Missouri" "Virginia" "Oklahoma" "California"
## [13] "Arizona" "New Mexico" "Illinois" "Kansas"
## [17] "Kentucky" "Nevada"
Reshaping data
Occasionally, you will have data in one arrangement (e.g. wide) that needs to be transformed into the long format. Perhaps this is needed for graphing purposes, or to compare particular data points, or because another R package requires it. Doing this manually is time consuming and error-prone. There are some function in tidyr that can do this very efficiently.
 Same data, two formats
Same data, two formats
Two common approaches to reshaping data:
- ‘tidyr’ (popular, fast)
- ‘data.table’ (flexible, very fast with large data sets)
| reshaping action | tidyr function | data.table function |
|---|---|---|
| long to wide | pivot_wider() |
dcast() |
| wide to long | pivot_longer() |
melt() |
Note: the tidyr functions pivot_wider() and pivot_longer() replaced spread() and gather().
The tidyr approach:
Reshaping from long to wide
# use the "barley_pnw" object created earlier (barley data from ID, OR and WA)
nass_barley_pnw_wide <- pivot_wider(barley_pnw,
id_cols = year,
names_from = state,
values_from = yield)
head(nass_barley_pnw_wide)
## # A tibble: 6 × 4
## year Oregon Idaho Washington
## <int> <dbl> <dbl> <dbl>
## 1 1869 35 NA NA
## 2 1870 30.5 NA NA
## 3 1871 28.5 NA NA
## 4 1872 30.5 NA NA
## 5 1873 33 NA NA
## 6 1874 29 NA NA
sample_n(nass_barley_pnw_wide, 5)
## # A tibble: 5 × 4
## year Oregon Idaho Washington
## <int> <dbl> <dbl> <dbl>
## 1 1869 35 NA NA
## 2 1911 23 35 32
## 3 1995 76 80 72
## 4 1956 37.5 33 35
## 5 1926 24 31 30.5
There is also a values_fill= argument to fill in missing values when you know what those should be. The default behavior is to fill with an NA.
Reshaping multiple variables
# use the "barley_pnw" object created earlier (barley data from ID, OR and WA)
nass_barley_pnw_wide2 <- pivot_wider(barley_pnw,
id_cols = year,
names_from = state,
values_from = c(yield, acres))
sample_n(nass_barley_pnw_wide2, 5)
## # A tibble: 5 × 7
## year yield_Oregon yield_Idaho yield_Washington acres_Oregon acres_I…¹ acres…²
## <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2004 73 92 70 66000 650000 245000
## 2 1945 29.5 37 35 257000 307000 125000
## 3 1960 36 28.5 36.5 457000 492000 661000
## 4 1914 23.5 32.5 33 82000 133000 187000
## 5 1946 34 34 37 278000 276000 90000
## # … with abbreviated variable names ¹acres_Idaho, ²acres_Washington
Reshaping from wide to long
nass_barley_pnw_long <- pivot_longer(nass_barley_pnw_wide,
cols = 2:ncol(nass_barley_pnw_wide),
names_to = "State",
values_to = "Yield",
values_drop_na = T)
head(nass_barley_pnw_long)
## # A tibble: 6 × 3
## year State Yield
## <int> <chr> <dbl>
## 1 1869 Oregon 35
## 2 1870 Oregon 30.5
## 3 1871 Oregon 28.5
## 4 1872 Oregon 30.5
## 5 1873 Oregon 33
## 6 1874 Oregon 29
dim(nass_barley_pnw_long)
## [1] 403 3
dim(barley_pnw) # dimension of the original object
## [1] 403 4
The data.table approach
Thus far, the package ‘data.table’ has not been explored in any depth (and it still won’t be for space considerations). Nevertheless, ‘data.table’ is useful for manipulating large tables; it is extremely efficient (even better than tidyverse functions).
Reshaping from long to wide:
-we will use the previously created data set “cotton_barley_inner” from inner join of the barley and cotton data sets.
# ID variable needs to be separated into State and year
cotton_barley_inner2 <- separate(cotton_barley_inner, ID, c("State", "Year"), sep = "_")
head(cotton_barley_inner2)
## # A tibble: 6 × 6
## State Year cotton_acres cotton_yield barley_acres barley_yield
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Tennessee 1866 649000 121 4000 23
## 2 Texas 1866 490000 325 3000 16
## 3 Tennessee 1867 611000 137 4000 19
## 4 Texas 1867 491000 195 2000 20
## 5 Tennessee 1868 520000 170 4000 18
## 6 Texas 1868 581000 197 2000 22
Reshape cotton acreage, using State as the ID variable:
cotton_acres_wide <- dcast(setDT(cotton_barley_inner2), State ~ Year, value.var = "cotton_acres")
Reshaping with multiple variables
Multiple ID variables can be specified on the left side of the tilde and/or multiple column categories can be specified on right side. Furthermore, multiple columns (the “value.var”) can be specified to fill the cells.
Reshape cotton acreage, using State as the ID variable using both cotton acres and cotton yield to fill the cells:
cotton_wide <- dcast(setDT(cotton_barley_inner2), State ~ Year, value.var = c("cotton_acres", "cotton_yield"))
sample(colnames(cotton_wide), 30)
## [1] "cotton_acres_1971" "cotton_acres_1898" "cotton_yield_1998"
## [4] "cotton_acres_1945" "cotton_yield_1947" "cotton_yield_1911"
## [7] "cotton_acres_1952" "cotton_yield_1944" "cotton_acres_1896"
## [10] "cotton_yield_1965" "cotton_yield_1945" "cotton_yield_1970"
## [13] "cotton_yield_2005" "cotton_yield_1957" "cotton_yield_1967"
## [16] "cotton_acres_1954" "cotton_yield_1898" "cotton_acres_1963"
## [19] "cotton_yield_1932" "cotton_acres_1926" "cotton_yield_1868"
## [22] "cotton_acres_1979" "cotton_yield_1908" "cotton_acres_1991"
## [25] "cotton_acres_1972" "cotton_acres_1878" "cotton_acres_1883"
## [28] "cotton_acres_1918" "cotton_yield_1921" "cotton_yield_1879"
Implementing a summary function in the reshape
- if there is more than one observation for each cell, a summary function like mean can be used. If function is not specified, the number of observations for that variable combination will be used.
cotton_wide_meanYield <- dcast(setDT(cotton_barley_inner2), cotton_acres ~ Year, value.var = "cotton_yield", fun = mean, fill = 0)
sample_n(cotton_wide_meanYield , 5)[,c(1, 100:106)]
## cotton_acres 1964 1965 1966 1967 1968 1969 1970
## 1: 571000 0 0 0 0 0 0 0
## 2: 420000 0 0 0 0 0 0 0
## 3: 3242000 0 0 0 0 0 0 0
## 4: 363000 0 0 0 0 0 0 0
## 5: 10212000 0 0 0 0 0 0 0
Reshaping from wide to long
cotton_acres_long <- melt(setDT(cotton_acres_wide),
id.vars = "State",
measure.vars = 2:ncol(cotton_acres_wide),
variable.name = "YEAR",
value.name = "ACRES")
sample_n(cotton_acres_long, 5)
## State YEAR ACRES
## 1: Nevada 1969 2300
## 2: Kentucky 1981 NA
## 3: Arkansas 1905 NA
## 4: Arkansas 1921 NA
## 5: Arkansas 1965 1205000
cotton_long <- melt(setDT(cotton_wide),
id.vars = "State",
measure.vars = list(2:147,148:ncol(cotton_wide)),
variable.name = "YEAR",
value.name = c("ACRES", "YIELD"), na.rm = T)
sample_n(cotton_long, 5)
## State YEAR ACRES YIELD
## 1: Kentucky 106 4300 573
## 2: Kansas 130 2600 185
## 3: New Mexico 72 159000 490
## 4: North Carolina 74 737000 296
## 5: Missouri 93 295000 446
Grouping and summarising data
- The function
group_by()provides a handy tool for breaking data into groups based on a common factor. - The function
summarise()often follows a grouping command. It will apply a summary function to a data set or a grouped data set. - these all rely on the idea of piping, or chaining together operations, using the pipe operator:
%>%, rather than (1) a complicated nesting arrangement (see below) or (2) a large number of intermediate objects that you don’t actually need.

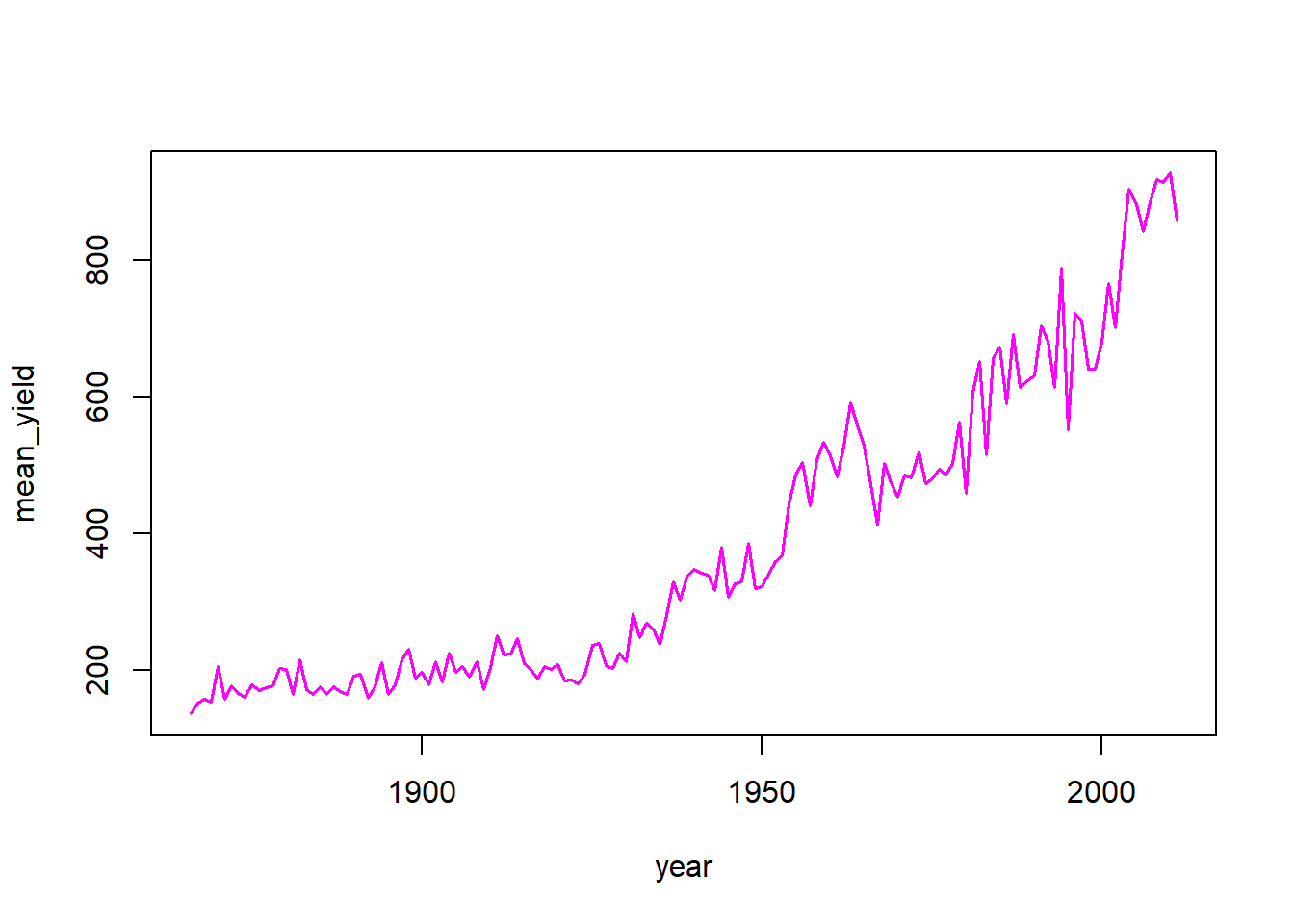
Example: group the cotton data set by year and output the mean yield for each year.
cotton_annual_mean <- group_by(nass_cotton, year) %>% summarise(mean_yield = mean(yield, na.rm = T))
with(cotton_annual_mean, plot(year, mean_yield, type = "l", col = "magenta", lwd = 1.5))
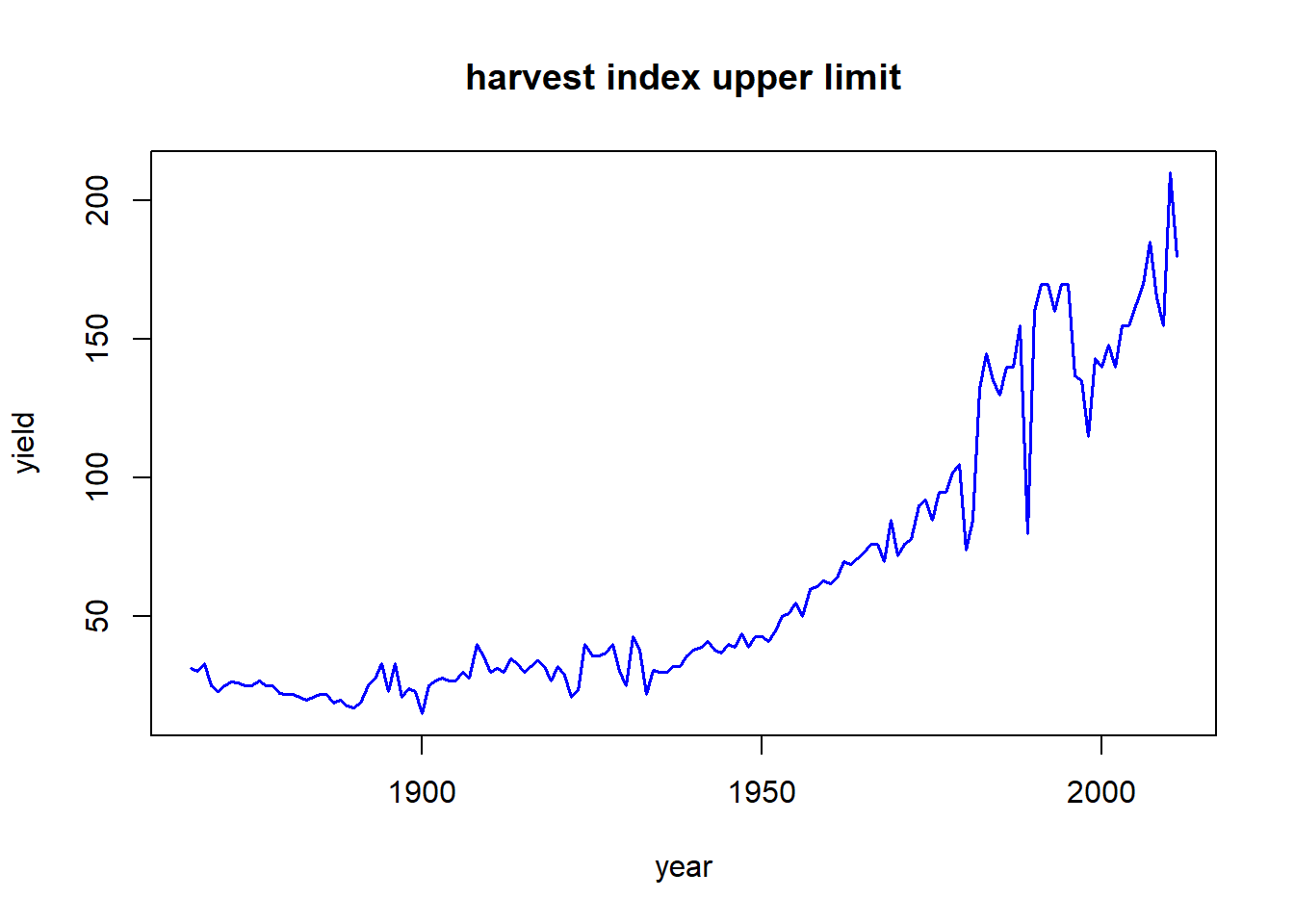
Example: Determine which states had the highest yields each year and plot highest yields over time
corn_highest_yielders <- nass_corn %>% mutate(HI = yield/acres) %>%
group_by(year) %>% slice(which.max(HI)) %>%
select(year, state, yield, HI) %>% mutate(state = as.character(state))
corn_highest_yielders %>% group_by(state) %>% tally(sort = T)
## # A tibble: 11 × 2
## state n
## <chr> <int>
## 1 Montana 27
## 2 Arizona 25
## 3 Nevada 24
## 4 Oregon 20
## 5 Rhode Island 20
## 6 Vermont 12
## 7 Utah 8
## 8 Maine 5
## 9 Wyoming 3
## 10 Connecticut 1
## 11 Idaho 1
with(corn_highest_yielders, plot(year, yield, type = "l", col = "blue", lwd = 1.5, main = "harvest index upper limit"))
Other Useful Functions
Not every amazing function in dplyr and tidyr are covered in this tutorial. Here are a few other functions you may find helpful when preparing and analyzing data.
| Package | Function | Purpose |
|---|---|---|
| dplyr | pull() |
extracts a single column and returns a vector |
| dplyr | n() |
counts observations |
| dplyr | n_distinct() |
counts the number of unique observations |
| dplyr | distinct() |
keeps only unique observations |
| dplyr | starts_with() |
used within select(), for selecting columns that start with a specific string of characters |
| tidyr | drop_na() |
remove rows with any missing data |
| tidyr | fill() |
fills in missing values with whatever occurs before it (column-wise). Its behavior is dependent on row order |
| dplyr | relocate() |
to move a column from one position to another (very helpful when working with a very wide data set) |
| tibble | rownames_to_column | will place rownames as the first column in a tibble or data.frame |
| tidyr | replace_na() |
replace missing values with a user-defined value (only when is.na(a_scalar) evaluates to “TRUE”) |
Other Resources
- R for data science - Thee Book for working with the tidyverse
- RStudio Cheat sheets - very handy!
- Package documentation - always worth checking out
- Tutorials by package authors: (these can often reveal package tricks that are hard to spot in the original help files)
vignette("dplyr")
